The Human RecQ4 Helicase Contains a Functional RecQ C-terminal Region (RQC) That Is Essential for Activity
- PMID: 27998982
- PMCID: PMC5354486
- DOI: 10.1074/jbc.M116.767954
The Human RecQ4 Helicase Contains a Functional RecQ C-terminal Region (RQC) That Is Essential for Activity
Abstract
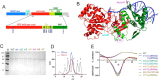
RecQ helicases are essential in the maintenance of genome stability. Five paralogues (RecQ1, Bloom, Werner, RecQ4, and RecQ5) are found in human cells, with distinct but overlapping roles. Mutations in human RecQ4 give rise to three distinct genetic disorders (Rothmund-Thomson, RAPADILINO, and Baller-Gerold syndromes), characterized by genetic instability, growth deficiency, and predisposition to cancer. Previous studies suggested that RecQ4 was unique because it did not seem to contain a RecQ C-terminal region (RQC) found in the other RecQ paralogues; such a region consists of a zinc domain and a winged helix domain and plays an important role in enzyme activity. However, our recent bioinformatic analysis identified in RecQ4 a putative RQC. To experimentally confirm this hypothesis, we report the purification and characterization of the catalytic core of human RecQ4. Inductively coupled plasma-atomic emission spectrometry detected the unusual presence of two zinc clusters within the zinc domain, consistent with the bioinformatic prediction. Analysis of site-directed mutants, targeting key RQC residues (putative zinc ligands and the aromatic residue predicted to be at the tip of the winged helix β-hairpin), showed a decrease in DNA binding, unwinding, and annealing, as expected for a functional RQC domain. Low resolution structural information obtained by small angle X-ray scattering data suggests that RecQ4 interacts with DNA in a manner similar to RecQ1, whereas the winged helix domain may assume alternative conformations, as seen in the bacterial enzymes. These combined results experimentally confirm the presence of a functional RQC domain in human RecQ4.
Keywords: DNA helicase; DNA repair; mutagenesis in vitro; protein purification; protein-DNA interaction; recombinant protein expression; small angle X-ray scattering (SAXS); structural model.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
-
- Larsen N. B., and Hickson I. D. (2013) RecQ helicases: conserved guardians of genomic integrity. Adv. Exp. Med. Biol. 767, 161–184 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

