Inferring human microbial dynamics from temporal metagenomics data: Pitfalls and lessons
- PMID: 28000336
- PMCID: PMC12268963
- DOI: 10.1002/bies.201600188
Inferring human microbial dynamics from temporal metagenomics data: Pitfalls and lessons
Abstract
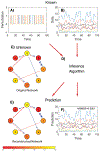
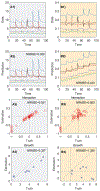
The human gut microbiota is a very complex and dynamic ecosystem that plays a crucial role in health and well-being. Inferring microbial community structure and dynamics directly from time-resolved metagenomics data is key to understanding the community ecology and predicting its temporal behavior. Many methods have been proposed to perform the inference. Yet, as we point out in this review, there are several pitfalls along the way. Indeed, the uninformative temporal measurements and the compositional nature of the relative abundance data raise serious challenges in inference. Moreover, the inference results can be largely distorted when only focusing on highly abundant species by ignoring or grouping low-abundance species. Finally, the implicit assumptions in various regularization methods may not reflect reality. Those issues have to be seriously considered in ecological modeling of human gut microbiota.
Keywords: dynamics inference; ecological modeling; human microbiome; temporal metagenomics.
© 2016 WILEY Periodicals, Inc.
Conflict of interest statement
The authors has declared no conflict of interest.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

