The balance of metagenomic elements shapes the skin microbiome in acne and health
- PMID: 28000755
- PMCID: PMC5175143
- DOI: 10.1038/srep39491
The balance of metagenomic elements shapes the skin microbiome in acne and health
Erratum in
-
Author Correction: The balance of metagenomic elements shapes the skin microbiome in acne and health.Sci Rep. 2020 Apr 2;10(1):6037. doi: 10.1038/s41598-020-62764-8. Sci Rep. 2020. PMID: 32242146 Free PMC article.
Abstract
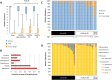
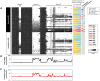
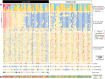
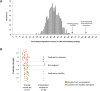
Studies have emphasized the importance of disease-associated microorganisms in perturbed communities, however, the protective roles of commensals are largely under recognized and poorly understood. Using acne as a model disease, we investigated the determinants of the overall virulence property of the skin microbiota when disease- and health-associated organisms coexist in the community. By ultra-deep metagenomic shotgun sequencing, we revealed higher relative abundances of propionibacteria and Propionibacterium acnes phage in healthy skin. In acne patients, the microbiome composition at the species level and at P. acnes strain level was more diverse than in healthy individuals, with enriched virulence-associated factors and reduced abundance of metabolic synthesis genes. Based on the abundance profiles of the metagenomic elements, we constructed a quantitative prediction model, which classified the clinical states of the host skin with high accuracy in both our study cohort (85%) and an independent sample set (86%). Our results suggest that the balance between metagenomic elements, not the mere presence of disease-associated strains, shapes the overall virulence property of the skin microbiota. This study provides new insights into the microbial mechanism of acne pathogenesis and suggests probiotic and phage therapies as potential acne treatments to modulate the skin microbiota and to maintain skin health.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

