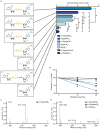
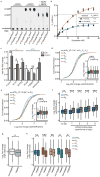
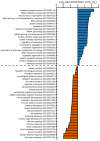
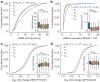
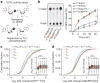
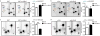Extended Data Figure 8. m6Am mRNAs are resistant to DCP2-mediated decapping and microRNA-mediated gene silencing
a, DCP2 decapping products are m7GDP. Here we confirm the identity of the putative m7GDP decapping product in the decapping assay by treatment with nucleoside-diphosphate kinase (NDPK). The shift to the m7GTP position confirms that the released product is m7GDP. A cap-labelled RNA with a guanosine as the first nucleotide was used as a positive control (lanes 3, 6, 9; the red ‘p’ denotes the position of the 32P). b, Michaelis-Menten curves of 10 nM DCP2 reacting with m7Gpppm6Am (blue) or m7GpppAm (orange) for 30 min at 37 °C. DCP2 shows higher decapping activity towards m7GpppAm than to m7Gpppm6Am (the dashed lines indicate the Km on the × axis; n = 3 biological replicates; mean ± s.e.m.). c, DCP2 depletion preferentially stabilizes Am mRNAs compared to m6Am mRNAs. Changes in half-life of Am mRNAs (FUCA1, PCK1, SCFD2) and m6Am mRNAs (PCNA, PSMD3, MAGOHB) in HEK293T cells transfected with either Flag vector (Ctrl) or DCP2-knockout cells (DCP2−/−) were determined by BrU pulse-chase analysis and subsequent qRT-PCR. Am mRNAs show a significant increase in half-life after DCP2 depletion whereas the half-life of m6Am mRNAs was not significantly increased. These data are related to the whole-transcriptome expression analysis presented in Fig. 4c and indicate that, in addition to the observed abundance changes of non-m6Am mRNAs versus m6Am mRNAs, DCP2 also selectively affects the half-life of specifically examined mRNAs (n = 3 biological replicates; mean ± s.e.m.; unpaired Student's t-test, *P ≤ 0.05, **P ≤ 0.01). d, In Fig. 4d, we found that m6Am mRNAs show less upregulation upon DICER knockdown than mRNAs beginning with other nucleotides. We wanted to further examine this concept using additional independent datasets of gene expression following depletion of proteins required for microRNA-mediated mRNA degradation, such as members of the Argonaute protein family. Measurement of mRNA expression in AGO2-knockdown HEK293T cells (siAGO2) compared to control cells (siCtrl) revealed more pronounced upregulation of non-m6Am mRNAs compared to those that have m6Am
(n = 2,080 (m6Am); 596 (Am); 1,085 (Cm); 805 (Gm); 1,274 (Um); data represent the average from two independent datasets; each box shows the first quartile, median, and third quartile; whiskers represent 1.5 × interquartile ranges; grey dots represent outliers; one-way ANOVA with Tukey's post hoc test, ***P ≤ 1 × 10−4 m6Am versus Am, Cm and Um). e, Similar to Fig. 4d, but here we only look at the expression changes of mRNAs that contain TargetScan-predicted microRNA-binding sites. Applying this filter criteria, we also observed that DICER knockdown in HEK293T cells (siDICER) resulted in more pronounced upregulation of non-m6Am miRNA target mRNAs compared to those that have m6Am
(n = 1,208 (m6Am); 359 (Am); 607 (Cm); 467 (Gm); 713 (Um); data represent the average from two independent datasets; each box shows the first quartile, median, and third quartile; whiskers represent 1.5 × interquartile ranges; grey dots represent outliers; one-way ANOVA with Tukey's post hoc test, ***P ≤ 9.6 × 10−4 versus m6AmNm, where Nm = Am, Cm, Gm or Um). f, In Fig. 4d and Extended Data Fig. 8e we show that m6Am mRNAs exhibit less upregulation upon DICER knockdown than mRNAs beginning with other nucleotides. We wanted to examine this concept further using additional filtering criteria. Thus, we asked if m6Am mRNA resistance to DICER depletion is dependent on the number of microRNA-binding sites. Therefore, we divided mRNAs into five groups: mRNAs that do not contain a predicted microRNA-binding site (0) and mRNAs that belong to specific quartiles that we assigned depending on the number of microRNA-binding sites (low (1) to high (4)). Notably, we did not observe any expression difference between m6Am mRNAs and non-m6Am mRNAs that do not carry predicted microRNA-binding sites. However, there was a clear increase in mRNA expression for mRNAs that contain microRNA-binding sites, and this increase was dependent on the number of microRNA-binding sites. Notably, for each quartile, m6Am mRNAs were significantly less upregulated than Nm mRNAs (n = 91 versus 89 (m6Am versus Nm; 1), 252 versus 339 (m6Am versus Nm; 1), 311 versus 454 (m6Am versus Nm; 2), 247 versus 541 (m6Am versus Nm; 3), 229 versus 512 (m6Am versus Nm; 4); data represent the average from two independent datasets; number of microRNA-binding sites in each quartile: 1 = 1–3; 2 = 4-6; 3 = 7–12; 4 = 13-54; each box shows the first quartile, median, and third quartile; whiskers represent 1.5 × interquartile ranges; one-way ANOVA with Tukey's post hoc test, *P ≤ 0.05, ***P ≤ 0.001, n.s., not significant). g, In Fig. 4d and Extended Data Fig. 9d-f we show that m6Am mRNAs are largely resistant to expression changes upon global inhibition of the microRNA machinery. We next asked whether introduction of a single microRNA also leads to differential responses of m6Am mRNAs compared to non-m6Am mRNAs. We used a published dataset where HeLa cells were transfected with a miR-155 duplex to achieve microRNA-specific mRNA degradation. For this analysis, we used m6Am mRNAs mapped in HEK293T cells. We first asked if we could observe a differential effect of mRNA degradation on miR-155 target and non-target mRNAs in the HeLa cell dataset. Indeed, miR-155 target mRNAs were significantly more suppressed in miR-155-transfected HeLa cells. This confirms that miR-155 target mRNA degradation can be detected in this dataset (n = 1,131 (target); 7,700 (non-target; data represent the average from two independent datasets; each box shows the first quartile, median, and third quartile; whiskers represent 1.5 × interquartile ranges; grey dots represent outliers; one-way ANOVA with Tukey's post hoc test, **P ≤ 2.2 × 10−16). h, m6Am mRNAs show resistance to miR-155-mediated mRNA degradation. We tested if the identity of the first nucleotide affects the response of miR-155 target mRNAs to miR-155-mediated mRNA degradation. We observed that miR-155 target mRNAs that start with m6Am show no significant suppression upon miR-155 transfection compared to non-target mRNAs that start with m6Am. However, expression of miR-155 target mRNAs that start with Am, Cm, Gm or Um was significantly suppressed compared to non-target mRNAs that start with Am, Cm, Gm or Um. These data suggest that the presence m6Am can reduce the silencing efficiency of a single microRNA in vivo (n = 1,714 versus 232 (m6Am, non-target versus target); 953 versus 158 (Am, non-target versus target); 1,848 versus 281 (Cm, non-target versus target); 1,394 versus 182 (Gm); 1,809 versus 278 (Um, non-target versus target); each box shows the first quartile, median, and third quartile; whiskers represent 1.5 × interquartile ranges; one-way ANOVA with Tukey's post hoc test, *P ≤ 0.05 non-target versus miR-155 target, **P ≤ 0.01 non-target versus miR-155 target, ***P ≤ 0.001 non-target versus miR-155 target).