Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity
- PMID: 28002404
- PMCID: PMC5570525
- DOI: 10.1038/nature20784
Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity
Abstract
Approximately 1.5 billion people worldwide are overweight or affected by obesity, and are at risk of developing type 2 diabetes, cardiovascular disease and related metabolic and inflammatory disturbances. Although the mechanisms linking adiposity to associated clinical conditions are poorly understood, recent studies suggest that adiposity may influence DNA methylation, a key regulator of gene expression and molecular phenotype. Here we use epigenome-wide association to show that body mass index (BMI; a key measure of adiposity) is associated with widespread changes in DNA methylation (187 genetic loci with P < 1 × 10-7, range P = 9.2 × 10-8 to 6.0 × 10-46; n = 10,261 samples). Genetic association analyses demonstrate that the alterations in DNA methylation are predominantly the consequence of adiposity, rather than the cause. We find that methylation loci are enriched for functional genomic features in multiple tissues (P < 0.05), and show that sentinel methylation markers identify gene expression signatures at 38 loci (P < 9.0 × 10-6, range P = 5.5 × 10-6 to 6.1 × 10-35, n = 1,785 samples). The methylation loci identify genes involved in lipid and lipoprotein metabolism, substrate transport and inflammatory pathways. Finally, we show that the disturbances in DNA methylation predict future development of type 2 diabetes (relative risk per 1 standard deviation increase in methylation risk score: 2.3 (2.07-2.56); P = 1.1 × 10-54). Our results provide new insights into the biologic pathways influenced by adiposity, and may enable development of new strategies for prediction and prevention of type 2 diabetes and other adverse clinical consequences of obesity.
Conflict of interest statement
None
Figures
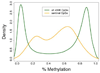

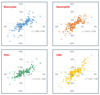


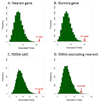
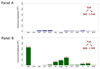

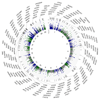

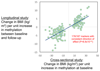

Comment in
-
Obesity: Methylation a consequence not a cause.Nat Rev Endocrinol. 2017 Mar;13(3):127. doi: 10.1038/nrendo.2016.223. Epub 2017 Jan 6. Nat Rev Endocrinol. 2017. PMID: 28059159 No abstract available.
References
MeSH terms
Substances
Grants and funding
- MR/K002414/1/MRC_/Medical Research Council/United Kingdom
- G0700931/MRC_/Medical Research Council/United Kingdom
- 084723/WT_/Wellcome Trust/United Kingdom
- G0601966/MRC_/Medical Research Council/United Kingdom
- PG/14/56/30976/BHF_/British Heart Foundation/United Kingdom
- MC_PC_15018/MRC_/Medical Research Council/United Kingdom
- MC_UU_12013/1/MRC_/Medical Research Council/United Kingdom
- RP-2015-06-005/DH_/Department of Health/United Kingdom
- RG/14/5/30893/BHF_/British Heart Foundation/United Kingdom
- MR/N015355/1/MRC_/Medical Research Council/United Kingdom
- MC_G0802523/MRC_/Medical Research Council/United Kingdom
- RP-PG-0407-10371/DH_/Department of Health/United Kingdom
- MC_UU_12013/8/MRC_/Medical Research Council/United Kingdom
- G9815508/MRC_/Medical Research Council/United Kingdom
- MC_UU_12013/2/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

