Sprouty2 correlates with favorable prognosis of gastric adenocarcinoma via suppressing FGFR2-induced ERK phosphorylation and cancer progression
- PMID: 28002800
- PMCID: PMC5354878
- DOI: 10.18632/oncotarget.13982
Sprouty2 correlates with favorable prognosis of gastric adenocarcinoma via suppressing FGFR2-induced ERK phosphorylation and cancer progression
Abstract
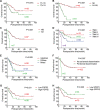
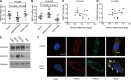
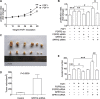
Fibroblast growth factor receptor 2 (FGFR2) has been identified as a predictive biomarker for unfavorable prognosis of gastric adenocarcinoma. As a well-defined antagonist in FGFR2-induced RAS/ERK activation, ectopic expression of sprouty (SPRY) family was reported in several kinds of cancers except gastric cancer. To explore the clinical significance of SPRY family and its correlation with FGFR2, we detected the expression of FGFR2 and SPRY family in 104 cases of gastric adenocarcinoma and subsequently analyzed their correlations with clinicopathological factors and overall survival rates by univariate and multivariate analysis. As the result, we demonstrated that both FGFR2 high-expression and SPRY2 low-expression indicated poorer prognosis of gastric adenocarcinoma. SPRY2 low-expression was significantly associated with FGFR2 high-expression, positive lymphatic invasion and metastasis. We further proved that SPRY2 could suppress FGFR2-induced ERK phosphorylation, cell proliferation and invasion with experiments in vitro and in vivo. In conclusion, we demonstrated that SPRY2 low-expression is a biomarker for unfavorable prognosis in gastric adenocarcinoma. SPRY2 can antagonize FGFR2-induced proliferation and invasion via suppressing ERK phosphorylation in gastric cancer cells, indicating SPRY2 as a potential therapeutic target for gastric adenocarcinoma treatment.
Keywords: FGFR2; SPRY2; gastric adenocarcinoma; invasion; proliferation.
Conflict of interest statement
The authors have no conflicts of interest.
Figures


References
-
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. - PubMed
-
- Torpy JM, Lynm C, Glass RM. JAMA patient page. Stomach cancer. JAMA. 2010;303:1771. - PubMed
-
- Brooks AN, Kilgour E, Smith PD. Molecular pathways: fibroblast growth factor signaling: a new therapeutic opportunity in cancer. Clin Cancer Res. 2012;18:1855–1862. - PubMed
-
- Katoh M, Nakagama H. FGF receptors: cancer biology and therapeutics. Med Res Rev. 2014;34:280–300. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

