Sub-lethal oxidative stress induces lysosome biogenesis via a lysosomal membrane permeabilization-cathepsin-caspase 3-transcription factor EB-dependent pathway
- PMID: 28002813
- PMCID: PMC5369955
- DOI: 10.18632/oncotarget.14016
Sub-lethal oxidative stress induces lysosome biogenesis via a lysosomal membrane permeabilization-cathepsin-caspase 3-transcription factor EB-dependent pathway
Abstract
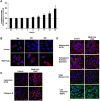
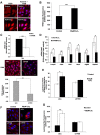
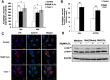
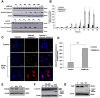
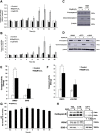
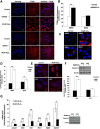
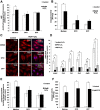
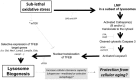
Here we provide evidence to link sub-lethal oxidative stress to lysosome biogenesis. Exposure of cells to sub-lethal concentrations of exogenously added hydrogen peroxide resulted in cytosol to nuclear translocation of the Transcription Factor EB (TFEB), the master controller of lysosome biogenesis and function. Nuclear translocation of TFEB was dependent upon the activation of a cathepsin-caspase 3 signaling pathway, downstream of lysosomal membrane permeabilization and accompanied by a significant increase in lysosome numbers as well as induction of TFEB-dependent lysosome-associated genes expression such as Ctsl, Lamp2 and its spliced variant Lamp2a, Neu1and Ctsb and Sqstm1 and Atg9b. The effects of sub-lethal oxidative stress on lysosomal gene expression and biogenesis were rescued upon gene silencing of caspase 3 and TFEB. Notably, caspase 3 activation was not associated with phenotypic hallmarks of apoptosis, evidenced by the absence of caspase 3 substrate cleavage, such as PARP, Lamin A/C or gelsolin. Taken together, these data demonstrate for the first time an unexpected and non-canonical role of a cathepsin-caspase 3 axis in the nuclear translocation of TFEB leading to lysosome biogenesis under conditions of sub-lethal oxidative stress.
Keywords: Autophagy; caspase 3; lysosomal membrane permeabilization; lysosomes; sub-lethal oxidative stress; transcription factor EB.
Conflict of interest statement
Authors declare that there is no conflict of interest.
Figures

Similar articles
-
Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration.Autophagy. 2019 Apr;15(4):631-651. doi: 10.1080/15548627.2018.1535292. Epub 2018 Nov 5. Autophagy. 2019. PMID: 30335591 Free PMC article.
-
The C-ETS2-TFEB Axis Promotes Neuron Survival under Oxidative Stress by Regulating Lysosome Activity.Oxid Med Cell Longev. 2016;2016:4693703. doi: 10.1155/2016/4693703. Epub 2016 Apr 19. Oxid Med Cell Longev. 2016. PMID: 27195074 Free PMC article.
-
Clomiphene citrate induces nuclear translocation of the TFEB transcription factor and triggers apoptosis by enhancing lysosomal membrane permeabilization.Biochem Pharmacol. 2019 Apr;162:191-201. doi: 10.1016/j.bcp.2018.11.016. Epub 2018 Nov 22. Biochem Pharmacol. 2019. PMID: 30471247
-
TFEB at a glance.J Cell Sci. 2016 Jul 1;129(13):2475-81. doi: 10.1242/jcs.146365. Epub 2016 Jun 1. J Cell Sci. 2016. PMID: 27252382 Free PMC article. Review.
-
Transcription factor EB: from master coordinator of lysosomal pathways to candidate therapeutic target in degenerative storage diseases.Ann N Y Acad Sci. 2016 May;1371(1):3-14. doi: 10.1111/nyas.13131. Ann N Y Acad Sci. 2016. PMID: 27299292 Free PMC article. Review.
Cited by
-
Past, present, and future perspectives of transcription factor EB (TFEB): mechanisms of regulation and association with disease.Cell Death Differ. 2022 Aug;29(8):1433-1449. doi: 10.1038/s41418-022-01028-6. Epub 2022 Jun 23. Cell Death Differ. 2022. PMID: 35739255 Free PMC article. Review.
-
Transcription factor EB as a key molecular factor in human health and its implication in diseases.SAGE Open Med. 2023 Feb 28;11:20503121231157209. doi: 10.1177/20503121231157209. eCollection 2023. SAGE Open Med. 2023. PMID: 36891126 Free PMC article. Review.
-
TFEB, a potential therapeutic target for osteoarthritis via autophagy regulation.Cell Death Dis. 2018 Aug 28;9(9):858. doi: 10.1038/s41419-018-0909-y. Cell Death Dis. 2018. PMID: 30154423 Free PMC article.
-
Lysosomal dysfunction-induced autophagic stress in diabetic kidney disease.J Cell Mol Med. 2020 Aug;24(15):8276-8290. doi: 10.1111/jcmm.15301. Epub 2020 Jun 25. J Cell Mol Med. 2020. PMID: 32583573 Free PMC article. Review.
-
MiT/TFE Family of Transcription Factors: An Evolutionary Perspective.Front Cell Dev Biol. 2021 Jan 6;8:609683. doi: 10.3389/fcell.2020.609683. eCollection 2020. Front Cell Dev Biol. 2021. PMID: 33490073 Free PMC article. Review.
References
-
- Davies KJ. The broad spectrum of responses to oxidants in proliferating cells: a new paradigm for oxidative stress. IUBMB Life. 1999;48(1):41–47. - PubMed
-
- Mates JM, Sanchez-Jimenez FM. Role of reactive oxygen species in apoptosis: implications for cancer therapy. Int J Biochem Cell Biol. 2000;32(2):157–170. - PubMed
-
- Jurk D, Wilson C, Passos JF, Oakley F, Correia-Melo C, Greaves L, Saretzki G, Fox C, Lawless C, Anderson R, Hewitt G, Pender SL, Fullard N, Nelson G, Mann J, van de Sluis B, et al. Chronic inflammation induces telomere dysfunction and accelerates ageing in mice. Nature communications. 2014;2:4172. - PMC - PubMed
-
- Starke-Reed PE, Oliver CN. Protein oxidation and proteolysis during aging and oxidative stress. Archives of biochemistry and biophysics. 1989;275(2):559–567. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

