Cholesterol metabolism: a potential therapeutic target in Mycobacteria
- PMID: 28002883
- PMCID: PMC5481656
- DOI: 10.1111/bph.13694
Cholesterol metabolism: a potential therapeutic target in Mycobacteria
Abstract
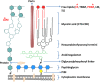
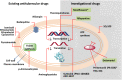
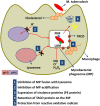
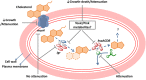
Tuberculosis (TB), although a curable disease, is still one of the most difficult infections to treat. Mycobacterium tuberculosis infects 10 million people worldwide and kills 1.5 million people each year. Reactivation of a latent infection is the major cause of TB. Cholesterol is a critical carbon source during latent infection. Catabolism of cholesterol contributes to the pool of propionyl-CoA, a precursor that is incorporated into lipid virulence factors. The M. tuberculosis genome contains a large regulon of cholesterol catabolic genes suggesting that the microorganism can utilize host sterol for infection and persistence. The protein products of these genes present ideal targets for rational drug discovery programmes. This review summarizes the development of enzyme inhibitors targeting the cholesterol pathway in M. tuberculosis. This knowledge is essential for the discovery of novel agents to treat M. tuberculosis infection.
Linked articles: This article is part of a themed section on Drug Metabolism and Antibiotic Resistance in Micro-organisms. To view the other articles in this section visit http://onlinelibrary.wiley.com/doi/10.1111/bph.v174.14/issuetoc.
© 2016 The British Pharmacological Society.
Figures


References
-
- Abuhammad A, Lack N, Schweichler J, Staunton D, Sim RB, Sim E (2011). Improvement of the expression and purification of Mycobacterium tuberculosis arylamine N‐acetyltransferase (TBNAT) a potential target for novel anti‐tubercular agents. Protein Expr Purif 80: 246–252. - PubMed
-
- Abuhammad A, Lowe ED, McDonough MA, Shaw Stewart PD, Kolek SA, Sim E et al. (2013). Structure of arylamine N‐acetyltransferase from Mycobacterium tuberculosis determined by cross‐seeding with the homologous protein from <styled-content style="fixed-case"><styled-content style="italic-in-any-context">M. marinum</styled-content></styled-content>: triumph over adversity. Acta Crystallogr D Biol Crystallogr 69: 1433–1446. - PubMed
-
- Abuhammad AM, Lowe ED, Fullam E, Noble M, Garman EF, Sim E (2010). Probing the architecture of the <styled-content style="fixed-case"><styled-content style="italic-in-any-context">Mycobacterium marinum</styled-content></styled-content> arylamine N‐acetyltransferase active site. Protein Cell 1: 384–392. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

