Fermentative Production of Cysteine by Pantoea ananatis
- PMID: 28003193
- PMCID: PMC5311407
- DOI: 10.1128/AEM.02502-16
Fermentative Production of Cysteine by Pantoea ananatis
Abstract
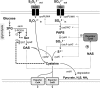
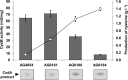
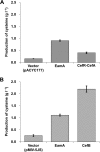
Cysteine is a commercially important amino acid; however, it lacks an efficient fermentative production method. Due to its cytotoxicity, intracellular cysteine levels are stringently controlled via several regulatory modes. Managing its toxic effects as well as understanding and deregulating the complexities of regulation are crucial for establishing the fermentative production of cysteine. The regulatory modes include feedback inhibition of key metabolic enzymes, degradation, efflux pumps, and the transcriptional regulation of biosynthetic genes by a master cysteine regulator, CysB. These processes have been extensively studied using Escherichia coli for overproducing cysteine by fermentation. In this study, we genetically engineered Pantoea ananatis, an emerging host for the fermentative production of bio-based materials, to identify key factors required for cysteine production. According to this and our previous studies, we identified a major cysteine desulfhydrase gene, ccdA (formerly PAJ_0331), involved in cysteine degradation, and the cysteine efflux pump genes cefA and cefB (formerly PAJ_3026 and PAJ_p0018, respectively), which may be responsible for downregulating the intracellular cysteine level. Our findings revealed that ccdA deletion and cefA and cefB overexpression are crucial factors for establishing fermentative cysteine production in P. ananatis and for obtaining a higher cysteine yield when combined with genes in the cysteine biosynthetic pathway. To our knowledge, this is the first demonstration of cysteine production in P. ananatis, which has fundamental implications for establishing overproduction in this microbe.IMPORTANCE The efficient production of cysteine is a major challenge in the amino acid fermentation industry. In this study, we identified cysteine efflux pumps and degradation pathways as essential elements and genetically engineered Pantoea ananatis, an emerging host for the fermentative production of bio-based materials, to establish the fermentative production of cysteine. This study provides crucial insights into the design and construction of cysteine-producing strains, which may play central roles in realizing commercial basis production.
Keywords: Pantoea ananatis; amino acid fermentation; cysteine; cysteine desulfhydrase; cysteine efflux.
Copyright © 2017 American Society for Microbiology.
Figures

References
-
- Hennicke F, Grumbt M, Lermann U, Ueberschaar N, Palige K, Bottcher B, Jacobsen ID, Staib C, Morschhauser J, Monod M, Hube B, Hertweck C, Staib P. 2013. Factors supporting cysteine tolerance and sulfite production in Candida albicans. Eukaryot Cell 12:604–613. doi:10.1128/EC.00336-12. - DOI - PMC - PubMed
-
- Kredich NM. 1996. Biosynthesis of cysteine, p 514–527. In Neidhardt FC, Curtis R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

