Blood Gene Signatures of Chagas Cardiomyopathy With or Without Ventricular Dysfunction
- PMID: 28003350
- PMCID: PMC6095079
- DOI: 10.1093/infdis/jiw540
Blood Gene Signatures of Chagas Cardiomyopathy With or Without Ventricular Dysfunction
Erratum in
-
Corrigendum to: Blood Gene Signatures of Chagas Cardiomyopathy With or Without Ventricular Dysfunction.J Infect Dis. 2020 Apr 7;221(9):1564. doi: 10.1093/infdis/jiz661. J Infect Dis. 2020. PMID: 31884515 Free PMC article. No abstract available.
Abstract
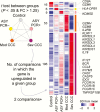
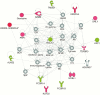
Chagas disease, caused by the protozoan parasite Trypanosoma cruzi, affects 7 million people in Latin American areas of endemicity. About 30% of infected patients will develop chronic Chagas cardiomyopathy (CCC), an inflammatory cardiomyopathy characterized by hypertrophy, fibrosis, and myocarditis. Further studies are necessary to understand the molecular mechanisms of disease progression. Transcriptome analysis has been increasingly used to identify molecular changes associated with disease outcomes. We thus assessed the whole-blood transcriptome of patients with Chagas disease. Microarray analysis was performed on blood samples from 150 subjects, of whom 30 were uninfected control patients and 120 had Chagas disease (1 group had asymptomatic disease, and 2 groups had CCC with either a preserved or reduced left ventricular ejection fraction [LVEF]). Each Chagas disease group displayed distinct gene expression and functional pathway profiles. The most different expression patterns were between CCC groups with a preserved or reduced LVEF. A more stringent analysis indicated that 27 differentially expressed genes, particularly those related to natural killer (NK)/CD8+ T-cell cytotoxicity, separated the 2 groups. NK/CD8+ T-cell cytotoxicity could play a role in determining Chagas disease progression. Understanding genes associated with disease may lead to improved insight into CCC pathogenesis and the identification of prognostic factors for CCC progression.
Keywords: Cardiomyopathy; Chagas disease; NK cells.; whole-blood transcriptome.
© The Author 2016. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures


References
-
- Morillo CA, Marin-Neto JA, Avezum A, et al. ; BENEFIT investigators Randomized trial of benznidazole for chronic Chagas’ cardiomyopathy. N Engl J Med 2015; 373:1295–306. - PubMed
-
- Gascon J, Vilasanjuan R, Lucas A. The need for global collaboration to tackle hidden public health crisis of Chagas disease. Expert Rev Anti Infect Ther 2014; 12:393–5. - PubMed
-
- Kalil-Filho R. Globalization of Chagas disease burden and new treatment perspectives. J Am Coll Cardiol 2015; 66:1190–2. - PubMed
-
- Cunha-Neto E, Bilate AM, Hyland KV, Fonseca SG, Kalil J, Engman DM. Induction of cardiac autoimmunity in Chagas heart disease: a case for molecular mimicry. Autoimmunity 2006; 39:41–54. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

