Microbiome Selection Could Spur Next-Generation Plant Breeding Strategies
- PMID: 28003808
- PMCID: PMC5141590
- DOI: 10.3389/fmicb.2016.01971
Microbiome Selection Could Spur Next-Generation Plant Breeding Strategies
Abstract
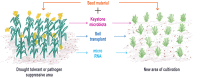
"No plant is an island too…" Plants, though sessile, have developed a unique strategy to counter biotic and abiotic stresses by symbiotically co-evolving with microorganisms and tapping into their genome for this purpose. Soil is the bank of microbial diversity from which a plant selectively sources its microbiome to suit its needs. Besides soil, seeds, which carry the genetic blueprint of plants during trans-generational propagation, are home to diverse microbiota that acts as the principal source of microbial inoculum in crop cultivation. Overall, a plant is ensconced both on the outside and inside with a diverse assemblage of microbiota. Together, the plant genome and the genes of the microbiota that the plant harbors in different plant tissues, i.e., the 'plant microbiome,' form the holobiome which is now considered as unit of selection: 'the holobiont.' The 'plant microbiome' not only helps plants to remain fit but also offers critical genetic variability, hitherto, not employed in the breeding strategy by plant breeders, who traditionally have exploited the genetic variability of the host for developing high yielding or disease tolerant or drought resistant varieties. This fresh knowledge of the microbiome, particularly of the rhizosphere, offering genetic variability to plants, opens up new horizons for breeding that could usher in cultivation of next-generation crops depending less on inorganic inputs, resistant to insect pest and diseases and resilient to climatic perturbations. We surmise, from ever increasing evidences, that plants and their microbial symbionts need to be co-propagated as life-long partners in future strategies for plant breeding. In this perspective, we propose bottom-up approach to co-propagate the co-evolved, the plant along with the target microbiome, through - (i) reciprocal soil transplantation method, or (ii) artificial ecosystem selection method of synthetic microbiome inocula, or (iii) by exploration of microRNA transfer method - for realizing this next-generation plant breeding approach. Our aim, thus, is to bring closer the information accrued through the advanced nucleotide sequencing and bioinformatics in conjunction with conventional culture-dependent isolation method for practical application in plant breeding and overall agriculture.
Keywords: artificial ecosystem selection; holobiont; microbiome; plant breeding; synthetic microbiota.
Figures

References
-
- Ambrosini A., de Souza R., Passaglia L. M. P. (2016). Ecological role of bacterial inoculants and their potential impact on soil microbial diversity. Plant Soil 400 193–207. 10.1007/s11104-015-2727-7 - DOI
-
- Badri D. V., Chaparro J. M., Zhang R., Shen Q., Vivanco J. M. (2013a). Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 288 4502–4512. 10.1074/jbc.A112.433300 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

