Assessing sedimentation equilibrium profiles in analytical ultracentrifugation experiments on macromolecules: from simple average molecular weight analysis to molecular weight distribution and interaction analysis
- PMID: 28003857
- PMCID: PMC5135724
- DOI: 10.1007/s12551-016-0232-8
Assessing sedimentation equilibrium profiles in analytical ultracentrifugation experiments on macromolecules: from simple average molecular weight analysis to molecular weight distribution and interaction analysis
Abstract
Molecular weights (molar masses), molecular weight distributions, dissociation constants and other interaction parameters are fundamental characteristics of proteins, nucleic acids, polysaccharides and glycoconjugates in solution. Sedimentation equilibrium analytical ultracentrifugation provides a powerful method with no supplementary immobilization, columns or membranes required. It is a particularly powerful tool when used in conjunction with its sister technique, namely sedimentation velocity. Here, we describe key approaches now available and their application to the characterization of antibodies, polysaccharides and glycoconjugates. We indicate how major complications, such as thermodynamic non-ideality, can now be routinely dealt with, thanks to a great extent to the extensive contribution of Professor Don Winzor over several decades of research.
Keywords: COVOL; Extended Fujita; Non-ideality; SEDFIT-MSTAR.
Conflict of interest statement
Compliance with ethical standards Conflict of interests Stephen E. Harding declares that he has no conflicts of interest. Richard B. Gillis declares that he has no conflicts of interest. Gary G. Adams declares that he has no conflicts of interest. Ethical approval This article does not contain any studies with human participants or animals performed by any of the authors.
Figures
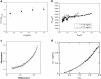
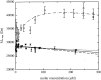
Similar articles
-
Application of novel analytical ultracentrifuge analysis to solutions of fungal mannans.Eur Biophys J. 2017 Apr;46(3):235-245. doi: 10.1007/s00249-016-1159-5. Epub 2016 Jul 21. Eur Biophys J. 2017. PMID: 27444285 Free PMC article.
-
Extended Fujita approach to the molecular weight distribution of polysaccharides and other polymeric systems.Methods. 2011 May;54(1):136-44. doi: 10.1016/j.ymeth.2011.01.009. Epub 2011 Jan 27. Methods. 2011. PMID: 21276851 Free PMC article.
-
Molecular weight distribution evaluation of polysaccharides and glycoconjugates using analytical ultracentrifugation.Macromol Biosci. 2010 Jul 7;10(7):714-20. doi: 10.1002/mabi.201000072. Macromol Biosci. 2010. PMID: 20632365 Review.
-
Insight into protein-protein interactions from analytical ultracentrifugation.Biochem Soc Trans. 2010 Aug;38(4):901-7. doi: 10.1042/BST0380901. Biochem Soc Trans. 2010. PMID: 20658974 Review.
-
SEDFIT-MSTAR: molecular weight and molecular weight distribution analysis of polymers by sedimentation equilibrium in the ultracentrifuge.Analyst. 2014 Jan 7;139(1):79-92. doi: 10.1039/c3an01507f. Epub 2013 Nov 18. Analyst. 2014. PMID: 24244936 Free PMC article.
Cited by
-
Optimizing high-throughput viral vector characterization with density gradient equilibrium analytical ultracentrifugation.Eur Biophys J. 2023 Jul;52(4-5):387-392. doi: 10.1007/s00249-023-01654-z. Epub 2023 May 2. Eur Biophys J. 2023. PMID: 37130969 Free PMC article.
-
Foreword to 'Quantitative and analytical relations in biochemistry'-a special issue in honour of Donald J. Winzor's 80th birthday.Biophys Rev. 2016 Dec;8(4):269-277. doi: 10.1007/s12551-016-0227-5. Epub 2016 Nov 4. Biophys Rev. 2016. PMID: 28510020 Free PMC article. Review.
-
Comparative Hydrodynamic Study on Non-Aqueous Soluble Archaeological Wood Consolidants: Butvar B-98 and PDMS-OH Siloxanes.Molecules. 2022 Mar 25;27(7):2133. doi: 10.3390/molecules27072133. Molecules. 2022. PMID: 35408530 Free PMC article.
-
Characterisation of mass distributions of solvent-fractionated lignins using analytical ultracentrifugation and size exclusion chromatography methods.Sci Rep. 2021 Jul 6;11(1):13937. doi: 10.1038/s41598-021-93424-0. Sci Rep. 2021. PMID: 34230572 Free PMC article.
-
Marine Algae Polysaccharides: An Overview of Characterization Techniques for Structural and Molecular Elucidation.Mar Drugs. 2025 Feb 27;23(3):105. doi: 10.3390/md23030105. Mar Drugs. 2025. PMID: 40137291 Free PMC article. Review.
References
-
- Cölfen H, Harding SE. MSTARA and MSTARI: interactive PC algorithms for simple, model independent evaluation of sedimentation equilibrium data. Eur Biophys J. 1997;25:333–346. doi: 10.1007/s002490050047. - DOI
-
- Cölfen H, Harding SE, Wilson EK, Scrutton NS, Winzor DJ. Low temperature solution behaviour of Methylophilus methylotrophus electron transferming flavoprotein: a study by analytical ultracentrifugation. Eur Biophys J. 1997;25:411–416. doi: 10.1007/s002490050054. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

