Transcriptome analysis reveals common differential and global gene expression profiles in bluetongue virus serotype 16 (BTV-16) infected peripheral blood mononuclear cells (PBMCs) in sheep and goats
- PMID: 28003963
- PMCID: PMC5157708
- DOI: 10.1016/j.gdata.2016.12.001
Transcriptome analysis reveals common differential and global gene expression profiles in bluetongue virus serotype 16 (BTV-16) infected peripheral blood mononuclear cells (PBMCs) in sheep and goats
Abstract
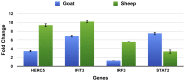
Bluetongue is an economically important infectious, arthropod borne viral disease of domestic and wild ruminants, caused by Bluetongue virus (BTV). Sheep are considered the most susceptible hosts, while cattle, buffalo and goats serve as reservoirs. The viral pathogenesis of BTV resulting in presence or absence of clinical disease among different hosts is not clearly understood. In the present study, transcriptome of sheep and goats peripheral blood mononuclear cells infected with BTV-16 was explored. The differentially expressed genes (DEGs) identified were found to be significantly enriched for immune system processes - NFκB signaling, MAPK signaling, Ras signaling, NOD signaling, RIG signaling, TNF signaling, TLR signaling, JAK-STAT signaling and VEGF signaling pathways. Greater numbers of DEGs were found to be involved in immune system processes in goats than in sheep. Interestingly, the DEHC (differentially expressed highly connected) gene network was found to be dense in goats than in sheep. Majority of the DEHC genes in the network were upregulated in goats but down-regulated in sheep. The network of differentially expressed immune genes with the other genes further confirmed these findings. Interferon stimulated genes - IFIT1 (ISG56), IFIT2 (ISG54) and IFIT3 (ISG60) responsible for antiviral state in the host were found to be upregulated in both the species. STAT2 was the TF commonly identified to co-regulate the DEGs, with its network showing genes that are downregulated in sheep but upregulated in goats. The genes dysregulated and the networks perturbed in the present study indicate host variability with a positive shift in immune response to BTV in goats than in sheep.
Keywords: Bluetongue; Bluetongue virus serotype-16; DEGs; DEHC genes; PBMCs; RNA-seq; transcriptome.
Figures

Similar articles
-
Transcriptome analysis reveals differential immune related genes expression in bovine viral diarrhea virus-2 infected goat peripheral blood mononuclear cells (PBMCs).BMC Genomics. 2019 Jun 21;20(1):516. doi: 10.1186/s12864-019-5830-y. BMC Genomics. 2019. PMID: 31226933 Free PMC article.
-
Comparative analysis of innate immune response following in vitro stimulation of sheep and goat peripheral blood mononuclear cells with bluetongue virus - serotype 23.Vet Res Commun. 2013 Dec;37(4):319-27. doi: 10.1007/s11259-013-9579-5. Epub 2013 Sep 22. Vet Res Commun. 2013. PMID: 24057859
-
Long-term infection of goats with bluetongue virus serotype 25.Vet Microbiol. 2013 Sep 27;166(1-2):165-73. doi: 10.1016/j.vetmic.2013.06.001. Epub 2013 Jun 18. Vet Microbiol. 2013. PMID: 23834964
-
Interferon α/β receptor knockout mice as a model to study bluetongue virus infection.Virus Res. 2014 Mar;182:35-42. doi: 10.1016/j.virusres.2013.09.038. Epub 2013 Oct 4. Virus Res. 2014. PMID: 24100234 Review.
-
Vaccines for Prevention of Bluetongue and Epizootic Hemorrhagic Disease in Livestock: A North American Perspective.Vector Borne Zoonotic Dis. 2015 Jun;15(6):385-96. doi: 10.1089/vbz.2014.1698. Vector Borne Zoonotic Dis. 2015. PMID: 26086559 Review.
Cited by
-
Whole-transcriptome analyses of sheep embryonic testicular cells infected with the bluetongue virus.Front Immunol. 2022 Dec 1;13:1053059. doi: 10.3389/fimmu.2022.1053059. eCollection 2022. Front Immunol. 2022. PMID: 36532076 Free PMC article.
-
Serological and molecular prevalence study of bluetongue virus in small domestic ruminants in Morocco.Sci Rep. 2022 Nov 14;12(1):19448. doi: 10.1038/s41598-022-24067-y. Sci Rep. 2022. PMID: 36376352 Free PMC article.
-
RNA Sequencing (RNA-Seq) Based Transcriptome Analysis in Immune Response of Holstein Cattle to Killed Vaccine against Bovine Viral Diarrhea Virus Type I.Animals (Basel). 2020 Feb 21;10(2):344. doi: 10.3390/ani10020344. Animals (Basel). 2020. PMID: 32098229 Free PMC article.
-
RNAseq Analysis of Brown Adipose Tissue and Thyroid of Newborn Lambs Subjected to Short-Term Cold Exposure Reveals Signs of Early Whitening of Adipose Tissue.Metabolites. 2022 Oct 20;12(10):996. doi: 10.3390/metabo12100996. Metabolites. 2022. PMID: 36295898 Free PMC article.
-
Construction and Analysis of ceRNA Networks Reveal the Key Genes Associated with Bovine Herpesvirus Type 1 Infection.Infect Drug Resist. 2023 Aug 31;16:5729-5740. doi: 10.2147/IDR.S411034. eCollection 2023. Infect Drug Resist. 2023. PMID: 37670981 Free PMC article.
References
-
- Maclachlan N.J., Drew C.P., Darpel K.E., Worwa G. The pathology and pathogenesis of bluetongue. J. Comp. Pathol. 2009;141:1–16. - PubMed
-
- Osburn B.I., McGowan B., Heron B., Loomis E., Bushnell R., Stott J., Utterback W. Epizootiologic study of bluetongue: virologic and serologic results. Am. J. Vet. Res. 1981;42:884–887. - PubMed
-
- Howell P.G., Verwoerd D.W. Bluetongue virus. Virol. Monogr. Virusforschung Einzeldarstellungen. 1971;9:35–74. - PubMed
-
- Verwoerd D.W., Huismans H. Studies on the in vitro and the in vivo transcription of the bluetongue virus genome. Onderstepoort J. Vet. Res. 1972;39:185–191. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

