p120-catenin prevents multinucleation through control of MKLP1-dependent RhoA activity during cytokinesis
- PMID: 28004812
- PMCID: PMC5192218
- DOI: 10.1038/ncomms13874
p120-catenin prevents multinucleation through control of MKLP1-dependent RhoA activity during cytokinesis
Erratum in
-
Corrigendum: p120-catenin prevents multinucleation through control of MKLP1-dependent RhoA activity during cytokinesis.Nat Commun. 2017 Sep 6;8:16030. doi: 10.1038/ncomms16030. eCollection 2017. Nat Commun. 2017. PMID: 31305775 Free PMC article.
Abstract
Spatiotemporal activation of RhoA and actomyosin contraction underpins cellular adhesion and division. Loss of cell-cell adhesion and chromosomal instability are cardinal events that drive tumour progression. Here, we show that p120-catenin (p120) not only controls cell-cell adhesion, but also acts as a critical regulator of cytokinesis. We find that p120 regulates actomyosin contractility through concomitant binding to RhoA and the centralspindlin component MKLP1, independent of cadherin association. In anaphase, p120 is enriched at the cleavage furrow where it binds MKLP1 to spatially control RhoA GTPase cycling. Binding of p120 to MKLP1 during cytokinesis depends on the N-terminal coiled-coil domain of p120 isoform 1A. Importantly, clinical data show that loss of p120 expression is a common event in breast cancer that strongly correlates with multinucleation and adverse patient survival. In summary, our study identifies p120 loss as a driver event of chromosomal instability in cancer.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
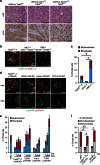
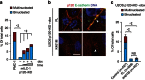

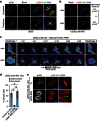

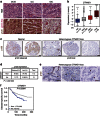
References
-
- Lengauer C., Kinzler K. W. & Vogelstein B. Genetic instability in colorectal cancers. Nature 386, 623–627 (1997). - PubMed
-
- Storchova Z. & Kuffer C. The consequences of tetraploidy and aneuploidy. J. Cell Sci. 121, 3859–3866 (2008). - PubMed
-
- Hernando E. et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430, 797–802 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

