Olaparib significantly delays photoreceptor loss in a model for hereditary retinal degeneration
- PMID: 28004814
- PMCID: PMC5177898
- DOI: 10.1038/srep39537
Olaparib significantly delays photoreceptor loss in a model for hereditary retinal degeneration
Abstract
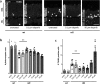
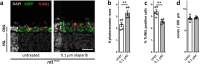
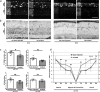
The enzyme poly-ADP-ribose-polymerase (PARP) mediates DNA-repair and rearrangements of the nuclear chromatin. Generally, PARP activity is thought to promote cell survival and in recent years a number of PARP inhibitors have been clinically developed for cancer treatment. Paradoxically, PARP activity is also connected to many diseases including the untreatable blinding disease Retinitis Pigmentosa (RP), where PARP activity appears to drive the pathogenesis of photoreceptor loss. We tested the efficacy of three different PARP inhibitors to prevent photoreceptor loss in the rd1 mouse model for RP. In retinal explant cultures in vitro, olaparib had strong and long-lasting photoreceptor neuroprotective capacities. We demonstrated target engagement by showing that olaparib reduced photoreceptor accumulation of poly-ADP-ribosylated proteins. Remarkably, olaparib also reduced accumulation of cyclic-guanosine-monophosphate (cGMP), a characteristic marker for photoreceptor degeneration. Moreover, intravitreal injection of olaparib in rd1 animals diminished PARP activity and increased photoreceptor survival, confirming in vivo neuroprotection. This study affirms the role of PARP in inherited retinal degeneration and for the first time shows that a clinically approved PARP inhibitor can prevent photoreceptor degeneration in an RP model. The wealth of human clinical data available for olaparib highlights its strong potential for a rapid clinical translation into a novel RP treatment.
Figures


Similar articles
-
Drug repurposing studies of PARP inhibitors as a new therapy for inherited retinal degeneration.Cell Mol Life Sci. 2020 Jun;77(11):2199-2216. doi: 10.1007/s00018-019-03283-2. Epub 2019 Aug 26. Cell Mol Life Sci. 2020. PMID: 31451894 Free PMC article.
-
PARP1 gene knock-out increases resistance to retinal degeneration without affecting retinal function.PLoS One. 2010 Nov 23;5(11):e15495. doi: 10.1371/journal.pone.0015495. PLoS One. 2010. PMID: 21124852 Free PMC article.
-
Excessive activation of poly(ADP-ribose) polymerase contributes to inherited photoreceptor degeneration in the retinal degeneration 1 mouse.J Neurosci. 2007 Sep 19;27(38):10311-9. doi: 10.1523/JNEUROSCI.1514-07.2007. J Neurosci. 2007. PMID: 17881537 Free PMC article.
-
Olaparib.Recent Results Cancer Res. 2018;211:217-233. doi: 10.1007/978-3-319-91442-8_15. Recent Results Cancer Res. 2018. PMID: 30069770 Review.
-
Olaparib: an oral PARP-1 and PARP-2 inhibitor with promising activity in ovarian cancer.Future Oncol. 2015;11(5):747-57. doi: 10.2217/fon.14.313. Future Oncol. 2015. PMID: 25757679 Review.
Cited by
-
Ceramide Induces the Death of Retina Photoreceptors Through Activation of Parthanatos.Mol Neurobiol. 2019 Jul;56(7):4760-4777. doi: 10.1007/s12035-018-1402-4. Epub 2018 Nov 1. Mol Neurobiol. 2019. PMID: 30387075
-
Determining Photoreceptor Cell Identity: Rod Versus Cone Fate Governed by tbx2b Opposing nrl.Invest Ophthalmol Vis Sci. 2024 Jan 2;65(1):39. doi: 10.1167/iovs.65.1.39. Invest Ophthalmol Vis Sci. 2024. PMID: 38261312 Free PMC article.
-
Reduced nuclear NAD+ drives DNA damage and subsequent immune activation in the retina.Hum Mol Genet. 2022 May 4;31(9):1370-1388. doi: 10.1093/hmg/ddab324. Hum Mol Genet. 2022. PMID: 34750622 Free PMC article.
-
Retinal Inflammation, Cell Death and Inherited Retinal Dystrophies.Int J Mol Sci. 2021 Feb 20;22(4):2096. doi: 10.3390/ijms22042096. Int J Mol Sci. 2021. PMID: 33672611 Free PMC article. Review.
-
Poly(ADP-ribose) polymerase inhibition: past, present and future.Nat Rev Drug Discov. 2020 Oct;19(10):711-736. doi: 10.1038/s41573-020-0076-6. Epub 2020 Sep 3. Nat Rev Drug Discov. 2020. PMID: 32884152 Review.
References
-
- De Vos M., Schreiber V. & Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem. Pharmacol. 84, 137–146 (2012). - PubMed
-
- Dryja T. P., Rucinski D. E., Chen S. H. & Berson E. L. Frequency of mutations in the gene encoding the alpha subunit of rod cGMP-phosphodiesterase in autosomal recessive retinitis pigmentosa. Invest Ophthalmol. Vis. Sci. 40, 1859–1865 (1999). - PubMed
-
- McLaughlin M. E., Sandberg M. A., Berson E. L. & Dryja T. P. Recessive mutations in the gene encoding the beta-subunit of rod phosphodiesterase in patients with retinitis pigmentosa. Nat. Genet. 4, 130–134 (1993). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

