A combined reference panel from the 1000 Genomes and UK10K projects improved rare variant imputation in European and Chinese samples
- PMID: 28004816
- PMCID: PMC5177868
- DOI: 10.1038/srep39313
A combined reference panel from the 1000 Genomes and UK10K projects improved rare variant imputation in European and Chinese samples
Abstract
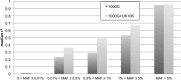
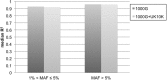
Imputation using the 1000 Genomes haplotype reference panel has been widely adapted to estimate genotypes in genome wide association studies. To evaluate imputation quality with a relatively larger reference panel and a reference panel composed of different ethnic populations, we conducted imputations in the Framingham Heart Study and the North Chinese Study using a combined reference panel from the 1000 Genomes (N = 1,092) and UK10K (N = 3,781) projects. For rare variants with 0.01% < MAF ≤ 0.5%, imputation in the Framingham Heart Study with the combined reference panel increased well-imputed genotypes (with imputation quality score ≥0.4) from 62.9% to 76.1% when compared to imputation with the 1000 Genomes. For the North Chinese samples, imputation of rare variants with 0.01% < MAF ≤ 0.5% with the combined reference panel increased well-imputed genotypes by from 49.8% to 61.8%. The predominant European ancestry of the UK10K and the combined reference panels may explain why there was less of an increase in imputation success in the North Chinese samples. Our results underscore the importance and potential of larger reference panels to impute rare variants, while recognizing that increasing ethnic specific variants in reference panels may result in better imputation for genotypes in some ethnic groups.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

