New CRISPR-Cas systems from uncultivated microbes
- PMID: 28005056
- PMCID: PMC5300952
- DOI: 10.1038/nature21059
New CRISPR-Cas systems from uncultivated microbes
Abstract
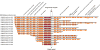
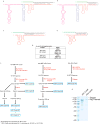
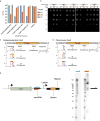
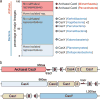
CRISPR-Cas systems provide microbes with adaptive immunity by employing short DNA sequences, termed spacers, that guide Cas proteins to cleave foreign DNA. Class 2 CRISPR-Cas systems are streamlined versions, in which a single RNA-bound Cas protein recognizes and cleaves target sequences. The programmable nature of these minimal systems has enabled researchers to repurpose them into a versatile technology that is broadly revolutionizing biological and clinical research. However, current CRISPR-Cas technologies are based solely on systems from isolated bacteria, leaving the vast majority of enzymes from organisms that have not been cultured untapped. Metagenomics, the sequencing of DNA extracted directly from natural microbial communities, provides access to the genetic material of a huge array of uncultivated organisms. Here, using genome-resolved metagenomics, we identify a number of CRISPR-Cas systems, including the first reported Cas9 in the archaeal domain of life, to our knowledge. This divergent Cas9 protein was found in little-studied nanoarchaea as part of an active CRISPR-Cas system. In bacteria, we discovered two previously unknown systems, CRISPR-CasX and CRISPR-CasY, which are among the most compact systems yet discovered. Notably, all required functional components were identified by metagenomics, enabling validation of robust in vivo RNA-guided DNA interference activity in Escherichia coli. Interrogation of environmental microbial communities combined with in vivo experiments allows us to access an unprecedented diversity of genomes, the content of which will expand the repertoire of microbe-based biotechnologies.
Conflict of interest statement
The Regents of the University of California have filed a provisional patent application related to the technology described in this work to the United States Patent and Trademark Office, in which D.B., L.B.H., S.C.S., J.A.D. and J.F.B. are listed as inventors.
Figures






Comment in
-
Metagenomics: Uncultivated microbes reveal new CRISPR-Cas systems.Nat Rev Genet. 2017 Mar;18(3):146. doi: 10.1038/nrg.2017.1. Epub 2017 Jan 23. Nat Rev Genet. 2017. PMID: 28111470 No abstract available.
-
New CRISPR-Cas systems discovered.Cell Res. 2017 Mar;27(3):313-314. doi: 10.1038/cr.2017.21. Epub 2017 Feb 21. Cell Res. 2017. PMID: 28220773 Free PMC article.
References
-
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
-
- Sorek R, Kunin V, Hugenholtz P. CRISPR — a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. - PubMed
-
- Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933–941. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

