Identification of Crucial Amino Acid Residues Involved in Agonist Signaling at the GPR55 Receptor
- PMID: 28005346
- PMCID: PMC5338039
- DOI: 10.1021/acs.biochem.6b01013
Identification of Crucial Amino Acid Residues Involved in Agonist Signaling at the GPR55 Receptor
Abstract
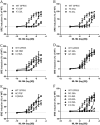
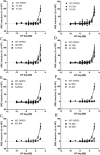
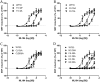
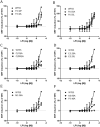
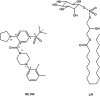
GPR55 is a newly deorphanized class A G-protein-coupled receptor that has been implicated in inflammatory pain, neuropathic pain, metabolic disorder, bone development, and cancer. Few potent GPR55 ligands have been identified to date. This is largely due to an absence of information about salient features of GPR55, such as residues important for signaling and residues implicated in the GPR55 signaling cascade. The goal of this work was to identify residues that are key for the signaling of the GPR55 endogenous ligand, l-α-lysophosphatidylinositol (LPI), as well as the signaling of the GPR55 agonist, ML184 {CID 2440433, 3-[4-(2,3-dimethylphenyl)piperazine-1-carbonyl]-N,N-dimethyl-4-pyrrolidin-1-ylbenzenesulfonamide}. Serum response element (SRE) and serum response factor (SRF) luciferase assays were used as readouts for studying LPI and ML184 signaling at the GPR55 mutants. A GPR55 R* model based on the recent δ-opioid receptor (DOR) crystal structure was used to interpret the resultant mutation data. Two residues were found to be crucial for agonist signaling at GPR55, K2.60 and E3.29, suggesting that these residues form the primary interaction site for ML184 and LPI at GPR55. Y3.32F, H(170)F, and F6.55A/L mutation results suggested that these residues are part of the orthosteric binding site for ML184, while Y3.32F and H(170)F mutation results suggest that these two residues are part of the LPI binding pocket. Y3.32L, M3.36A, and F6.48A mutation results suggest the importance of a Y3.32/M3.36/F6.48 cluster in the GPR55 signaling cascade. C(10)A and C(260)A mutations suggest that these residues form a second disulfide bridge in the extracellular domain of GPR55, occluding ligand extracellular entry in the TMH1-TMH7 region of GPR55. Taken together, these results provide the first set of discrete information about GPR55 residues important for LPI and ML184 signaling and for GPR55 activation. This information should aid in the rational design of next-generation GPR55 ligands and the creation of the first high-affinity GPR55 radioligand, a tool that is sorely needed in the field.
Figures
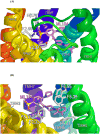



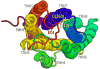


References
-
- Henstridge CM, Balenga NA, Ford LA, Ross RA, Waldhoer M, Irving AJ. The GPR55 ligand L-alpha-lysophosphatidylinositol promotes RhoA-dependent Ca2+ signaling and NFAT activation. Faseb J. 2009;23(1):183–93. - PubMed
- Oka S, Toshida T, Maruyama K, Nakajima K, Yamashita A, Sugiura T. 2-Arachidonoyl-sn-glycero-3-phosphoinositol: A Possible Natural Ligand for GPR55. Journal of biochemistry. 2009;145(1):13–20. - PubMed
- Oka S, Nakajima K, Yamashita A, Kishimoto S, Sugiura T. Identification of GPR55 as a lysophosphatidylinositol receptor. Biochem Biophys Res Commun. 2007;362(4):928–34. - PubMed
-
- Sawzdargo M, Nguyen T, Lee DK, Lynch KR, Cheng R, Heng HH, George SR, O'Dowd BF. Identification and cloning of three novel human G protein-coupled receptor genes GPR52, PsiGPR53 and GPR55: GPR55 is extensively expressed in human brain. Brain Res Mol Brain Res. 1999;64(2):193–8. - PubMed
- Staton PC, Hatcher JP, Walker DJ, Morrison AD, Shapland EM, Hughes JP, Chong E, Mander PK, Green PJ, Billinton A, Fulleylove M, Lancaster HC, Smith JC, Bailey LT, Wise A, Brown AJ, Richardson JC, Chessell IP. The putative cannabinoid receptor GPR55 plays a role in mechanical hyperalgesia associated with inflammatory and neuropathic pain. Pain. 2008;139(1):225–36. - PubMed
- Whyte LS, Ryberg E, Sims NA, Ridge SA, Mackie K, Greasley PJ, Ross RA, Rogers MJ. The putative cannabinoid receptor GPR55 affects osteoclast function in vitro and bone mass in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(38):16511–6. - PMC - PubMed
- Ford LA, Roelofs AJ, Anavi-Goffer S, Mowat L, Simpson DG, Irving AJ, Rogers MJ, Rajnicek AM, Ross RA. A role for L-alpha-lysophosphatidylinositol and GPR55 in the modulation of migration, orientation and polarization of human breast cancer cells. Br J Pharmacol. 2010;160(3):762–71. - PMC - PubMed
- Andradas C, Caffarel MM, Perez-Gomez E, Salazar M, Lorente M, Velasco G, Guzman M, Sanchez C. The orphan G protein-coupled receptor GPR55 promotes cancer cell proliferation via ERK. Oncogene. 2010;30:245–252. - PubMed
- Pineiro R, Maffucci T, Falasca M. The putative cannabinoid receptor GPR55 defines a novel autocrine loop in cancer cell proliferation. Oncogene. 2010;30:142–152. - PubMed
-
- Heynen-Genel S, Dahl R, Shi S, Milan L, Hariharan S, Sergienko E, Hedrick M, Dad S, Stonich D, Su Y, Vicchiarelli M, Mangravita-Novo A, Smith LH, Chung TDY, Sharir H, Caron MG, Barak LS, Abood ME. Screening for Selective Ligands for GPR55 - Antagonists. Probe Reports NIH Molecular Libraries Program. 2010 - PubMed
- Heynen-Genel S, Dahl R, Shi S, Milan L, Hariharan S, Bravo Y, Sergienko E, Hedrick M, Dad S, Stonich D, Su Y, Vicchiarelli M, Mangravita-Novo A, Smith LH, Chung TDY, Sharir H, Barak LS, Abood ME. Screening for Selective Ligands for GPR55 - Agonists. Probe Reports NIH Molecular Libraries Program. 2010 - PubMed
-
- Heynen-Genel S, Dahl R, Shi S, Milan L, Hariharan S, Bravo Y, Sergienko E, Hedrick M, Dad S, Stonich D, Su Y, Vicchiarelli M, Mangravita-Novo A, Smith LH, Chung TDY, Sharir H, Barak LS, Abood ME. Screening for Selective Ligands for GPR55. Probe Reports NIH Molecular Libraries Program. 2010
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

