Small GTPases controlling autophagy-related membrane traffic in yeast and metazoans
- PMID: 28005455
- PMCID: PMC6205004
- DOI: 10.1080/21541248.2016.1258444
Small GTPases controlling autophagy-related membrane traffic in yeast and metazoans
Abstract
During macroautophagy, the phagophore-mediated formation of autophagosomes and their subsequent fusion with lysosomes requires extensive transformation of the endomembrane system. Membrane dynamics in eukaryotic cells is regulated by small GTPase proteins including Arfs and Rabs. The small GTPase proteins that regulate autophagic membrane traffic are mostly conserved in yeast and metazoans, but there are also several differences. In this mini-review, we compare the small GTPase network of yeast and metazoan cells that regulates autophagy, and point out the similarities and differences in these organisms.
Keywords: Arf; Rab; Rag; TORC1; autolysosome; autophagosome; autophagy; membrane fusion; membrane trafficking.
Figures
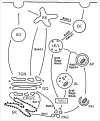
References
-
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069-75; PMID:18305538; http://dx.doi.org/ 10.1038/nature06639 - DOI - PMC - PubMed
-
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008; 320:1496-501; PMID:18497260; http://dx.doi.org/ 10.1126/science.1157535 - DOI - PMC - PubMed
-
- Kira S, Tabata K, Shirahama-Noda K, Nozoe A, Yoshimori T, Noda T. Reciprocal conversion of Gtr1 and Gtr2 nucleotide-binding states by Npr2-Npr3 inactivates TORC1 and induces autophagy. Autophagy 2014; 10:1565-78; PMID:25046117; http://dx.doi.org/ 10.4161/auto.29397 - DOI - PMC - PubMed
-
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 2008; 10:935-45; PMID:18604198; http://dx.doi.org/ 10.1038/ncb1753 - DOI - PMC - PubMed
-
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol 2010; 22:132-9; PMID:20056399; http://dx.doi.org/ 10.1016/j.ceb.2009.12.004 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
