Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies
- PMID: 28005526
- PMCID: PMC5563544
- DOI: 10.1099/ijsem.0.001755
Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies
Abstract
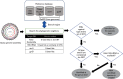
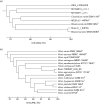
The recent advent of DNA sequencing technologies facilitates the use of genome sequencing data that provide means for more informative and precise classification and identification of members of the Bacteria and Archaea. Because the current species definition is based on the comparison of genome sequences between type and other strains in a given species, building a genome database with correct taxonomic information is of paramount need to enhance our efforts in exploring prokaryotic diversity and discovering novel species as well as for routine identifications. Here we introduce an integrated database, called EzBioCloud, that holds the taxonomic hierarchy of the Bacteria and Archaea, which is represented by quality-controlled 16S rRNA gene and genome sequences. Whole-genome assemblies in the NCBI Assembly Database were screened for low quality and subjected to a composite identification bioinformatics pipeline that employs gene-based searches followed by the calculation of average nucleotide identity. As a result, the database is made of 61 700 species/phylotypes, including 13 132 with validly published names, and 62 362 whole-genome assemblies that were identified taxonomically at the genus, species and subspecies levels. Genomic properties, such as genome size and DNA G+C content, and the occurrence in human microbiome data were calculated for each genus or higher taxa. This united database of taxonomy, 16S rRNA gene and genome sequences, with accompanying bioinformatics tools, should accelerate genome-based classification and identification of members of the Bacteria and Archaea. The database and related search tools are available at www.ezbiocloud.net/.
Conflict of interest statement
Authors are employees of ChunLab, Inc., a company that provides bioinformatics services in microbial genomics and metagenomics. ChunLab paid for the research.
Figures
Similar articles
-
EzBioCloud: a genome-driven database and platform for microbiome identification and discovery.Int J Syst Evol Microbiol. 2024 Jun;74(6):006421. doi: 10.1099/ijsem.0.006421. Int J Syst Evol Microbiol. 2024. PMID: 38888585 Free PMC article.
-
Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species.Int J Syst Evol Microbiol. 2012 Mar;62(Pt 3):716-721. doi: 10.1099/ijs.0.038075-0. Epub 2011 Nov 25. Int J Syst Evol Microbiol. 2012. PMID: 22140171
-
The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains.Syst Appl Microbiol. 2008 Sep;31(4):241-50. doi: 10.1016/j.syapm.2008.07.001. Epub 2008 Aug 9. Syst Appl Microbiol. 2008. PMID: 18692976
-
Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences.Nat Rev Microbiol. 2014 Sep;12(9):635-45. doi: 10.1038/nrmicro3330. Nat Rev Microbiol. 2014. PMID: 25118885 Review.
-
Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea.Int J Syst Evol Microbiol. 2014 Feb;64(Pt 2):316-324. doi: 10.1099/ijs.0.054171-0. Int J Syst Evol Microbiol. 2014. PMID: 24505069 Review.
Cited by
-
Complete Genome Sequence of Bacillus sp. Strain V3, Isolated from Mangrove Sediments in Wenzhou, China.Microbiol Resour Announc. 2021 May 6;10(18):e01448-20. doi: 10.1128/MRA.01448-20. Microbiol Resour Announc. 2021. PMID: 33958409 Free PMC article.
-
Cell Plasticity and Genomic Structure of a Novel Filterable Rhizobiales Bacterium that Belongs to a Widely Distributed Lineage.Microorganisms. 2020 Sep 7;8(9):1373. doi: 10.3390/microorganisms8091373. Microorganisms. 2020. PMID: 32906802 Free PMC article.
-
Bacteremia caused by Comamonas kerstersii in a patient with acute perforated appendicitis and localized peritonitis: case report and literature review.Front Med (Lausanne). 2023 Dec 7;10:1283769. doi: 10.3389/fmed.2023.1283769. eCollection 2023. Front Med (Lausanne). 2023. PMID: 38131046 Free PMC article.
-
Cultivation of Diverse Novel Marine Bacteria from Deep Ocean Sediment Using Spent Culture Supernatant of Ca. Bathyarchaeia Enrichment.J Microbiol. 2024 Aug;62(8):611-625. doi: 10.1007/s12275-024-00145-w. Epub 2024 Jul 10. J Microbiol. 2024. PMID: 38985432
-
Description of Mycobacterium pinniadriaticum sp. nov., isolated from a noble pen shell (Pinna nobilis) population in Croatia.Front Microbiol. 2023 Dec 15;14:1289182. doi: 10.3389/fmicb.2023.1289182. eCollection 2023. Front Microbiol. 2023. PMID: 38192290 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

