Drug Repositioning for Alzheimer's Disease Based on Systematic 'omics' Data Mining
- PMID: 28005991
- PMCID: PMC5179106
- DOI: 10.1371/journal.pone.0168812
Drug Repositioning for Alzheimer's Disease Based on Systematic 'omics' Data Mining
Abstract
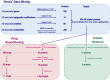
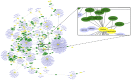
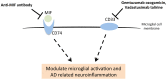
Traditional drug development for Alzheimer's disease (AD) is costly, time consuming and burdened by a very low success rate. An alternative strategy is drug repositioning, redirecting existing drugs for another disease. The large amount of biological data accumulated to date warrants a comprehensive investigation to better understand AD pathogenesis and facilitate the process of anti-AD drug repositioning. Hence, we generated a list of anti-AD protein targets by analyzing the most recent publically available 'omics' data, including genomics, epigenomics, proteomics and metabolomics data. The information related to AD pathogenesis was obtained from the OMIM and PubMed databases. Drug-target data was extracted from the DrugBank and Therapeutic Target Database. We generated a list of 524 AD-related proteins, 18 of which are targets for 75 existing drugs-novel candidates for repurposing as anti-AD treatments. We developed a ranking algorithm to prioritize the anti-AD targets, which revealed CD33 and MIF as the strongest candidates with seven existing drugs. We also found 7 drugs inhibiting a known anti-AD target (acetylcholinesterase) that may be repurposed for treating the cognitive symptoms of AD. The CAD protein and 8 proteins implicated by two 'omics' approaches (ABCA7, APOE, BIN1, PICALM, CELF1, INPP5D, SPON1, and SOD3) might also be promising targets for anti-AD drug development. Our systematic 'omics' mining suggested drugs with novel anti-AD indications, including drugs modulating the immune system or reducing neuroinflammation that are particularly promising for AD intervention. Furthermore, the list of 524 AD-related proteins could be useful not only as potential anti-AD targets but also considered for AD biomarker development.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Ghani M, Rogaeva E. Autosomal dominant Alzheimer’s disease: underlying causes In: Galimberti D, Scarpini E., editor. Neurodegenerative Diseases. London: Springer; 2014. p. 27–47.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

