iCAGES: integrated CAncer GEnome Score for comprehensively prioritizing driver genes in personal cancer genomes
- PMID: 28007024
- PMCID: PMC5180414
- DOI: 10.1186/s13073-016-0390-0
iCAGES: integrated CAncer GEnome Score for comprehensively prioritizing driver genes in personal cancer genomes
Abstract
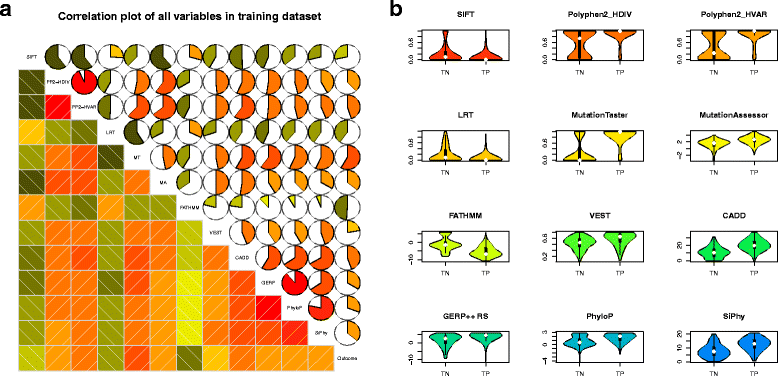
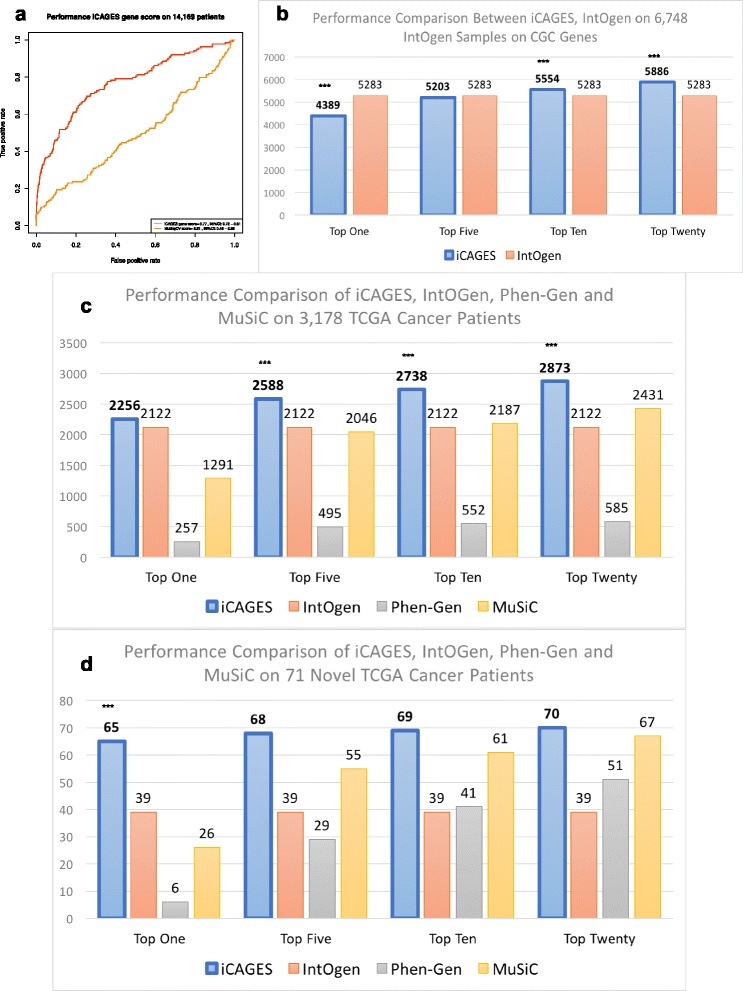
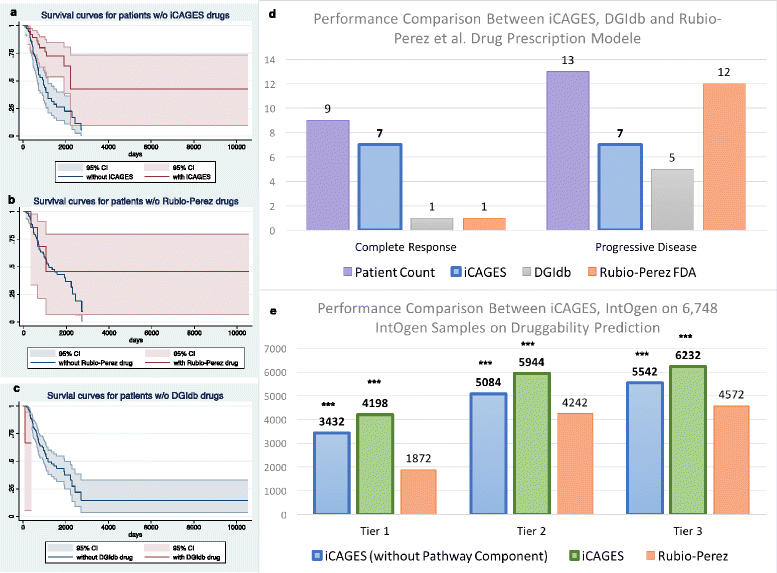
Cancer results from the acquisition of somatic driver mutations. Several computational tools can predict driver genes from population-scale genomic data, but tools for analyzing personal cancer genomes are underdeveloped. Here we developed iCAGES, a novel statistical framework that infers driver variants by integrating contributions from coding, non-coding, and structural variants, identifies driver genes by combining genomic information and prior biological knowledge, then generates prioritized drug treatment. Analysis on The Cancer Genome Atlas (TCGA) data showed that iCAGES predicts whether patients respond to drug treatment (P = 0.006 by Fisher's exact test) and long-term survival (P = 0.003 from Cox regression). iCAGES is available at http://icages.wglab.org .
Keywords: Cancer genomics; Machine learning; Precision medicine; Precision oncology; TCGA.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

