Genetics and Genomics of Single-Gene Cardiovascular Diseases: Common Hereditary Cardiomyopathies as Prototypes of Single-Gene Disorders
- PMID: 28007145
- PMCID: PMC5189923
- DOI: 10.1016/j.jacc.2016.09.968
Genetics and Genomics of Single-Gene Cardiovascular Diseases: Common Hereditary Cardiomyopathies as Prototypes of Single-Gene Disorders
Abstract
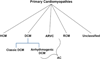
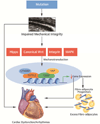
This is the first of 2 review papers on genetics and genomics appearing as part of the series on "omics." Genomics pertains to all components of an organism's genes, whereas genetics involves analysis of a specific gene or genes in the context of heredity. The paper provides introductory comments, describes the basis of human genetic diversity, and addresses the phenotypic consequences of genetic variants. Rare variants with large effect sizes are responsible for single-gene disorders, whereas complex polygenic diseases are typically due to multiple genetic variants, each exerting a modest effect size. To illustrate the clinical implications of genetic variants with large effect sizes, 3 common forms of hereditary cardiomyopathies are discussed as prototypic examples of single-gene disorders, including their genetics, clinical manifestations, pathogenesis, and treatment. The genetic basis of complex traits is discussed in a separate paper.
Keywords: cardiomyopathy; mutation; noncoding RNA.
Copyright © 2016 American College of Cardiology Foundation. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Reviews of translational medicine and genomics in cardiovascular disease: new disease taxonomy and therapeutic implications cardiomyopathies: therapeutics based on molecular phenotype.J Am Coll Cardiol. 2007 Mar 27;49(12):1251-64. doi: 10.1016/j.jacc.2006.10.073. Epub 2007 Mar 9. J Am Coll Cardiol. 2007. PMID: 17394955 Review.
-
New approaches to establish genetic causality.Trends Cardiovasc Med. 2015 Oct;25(7):646-52. doi: 10.1016/j.tcm.2015.02.013. Epub 2015 Mar 4. Trends Cardiovasc Med. 2015. PMID: 25864169 Free PMC article. Review.
-
Exploring predisposition and treatment response--the promise of genomics.Prog Cardiovasc Dis. 2012 Jul-Aug;55(1):56-63. doi: 10.1016/j.pcad.2012.04.006. Prog Cardiovasc Dis. 2012. PMID: 22824110 Free PMC article. Review.
-
Linking Genes to Cardiovascular Diseases: Gene Action and Gene-Environment Interactions.J Cardiovasc Transl Res. 2015 Dec;8(9):506-27. doi: 10.1007/s12265-015-9658-9. Epub 2015 Nov 6. J Cardiovasc Transl Res. 2015. PMID: 26545598 Free PMC article. Review.
-
Implications of genetic testing in noncompaction/hypertrabeculation.Am J Med Genet C Semin Med Genet. 2013 Aug;163C(3):206-11. doi: 10.1002/ajmg.c.31371. Epub 2013 Jul 10. Am J Med Genet C Semin Med Genet. 2013. PMID: 23843345 Free PMC article.
Cited by
-
Application of Spatial Omics in the Cardiovascular System.Research (Wash D C). 2025 Mar 8;8:0628. doi: 10.34133/research.0628. eCollection 2025. Research (Wash D C). 2025. PMID: 40062231 Free PMC article. Review.
-
Rationale and design of the PROspective ATHletic Heart (Pro@Heart) study: long-term assessment of the determinants of cardiac remodelling and its clinical consequences in endurance athletes.BMJ Open Sport Exerc Med. 2022 Mar 18;8(1):e001309. doi: 10.1136/bmjsem-2022-001309. eCollection 2022. BMJ Open Sport Exerc Med. 2022. PMID: 35368514 Free PMC article.
-
Distinct hypertrophic cardiomyopathy genotypes result in convergent sarcomeric proteoform profiles revealed by top-down proteomics.Proc Natl Acad Sci U S A. 2020 Oct 6;117(40):24691-24700. doi: 10.1073/pnas.2006764117. Epub 2020 Sep 23. Proc Natl Acad Sci U S A. 2020. PMID: 32968017 Free PMC article.
-
Genetic Pathogenesis of Hypertrophic and Dilated Cardiomyopathy.Heart Fail Clin. 2018 Apr;14(2):139-146. doi: 10.1016/j.hfc.2017.12.004. Heart Fail Clin. 2018. PMID: 29525643 Free PMC article. Review.
-
Unraveling obscurins in heart disease.Pflugers Arch. 2019 May;471(5):735-743. doi: 10.1007/s00424-018-2191-3. Epub 2018 Aug 11. Pflugers Arch. 2019. PMID: 30099631 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

