Small Cell Lung Cancer Exhibits Frequent Inactivating Mutations in the Histone Methyltransferase KMT2D/MLL2: CALGB 151111 (Alliance)
- PMID: 28007623
- PMCID: PMC5669801
- DOI: 10.1016/j.jtho.2016.12.011
Small Cell Lung Cancer Exhibits Frequent Inactivating Mutations in the Histone Methyltransferase KMT2D/MLL2: CALGB 151111 (Alliance)
Abstract
Introduction: SCLC is a lethal neuroendocrine tumor type that is highly prone to metastasis. There is an urgency to understand the mutated genes that promote SCLC, as there are no approved targeted therapies yet available. SCLC is rarely resected, limiting the number of samples available for genomic analyses of somatic mutations.
Methods: To identify potential driver mutations in human SCLC we sequenced the whole exomes of 18 primary SCLCs and seven cell lines along with matched normal controls. We extended these data by resequencing a panel of genes across 40 primary SCLCs and 48 cell lines.
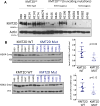
Results: We report frequent mutations in the lysine methyltransferase 2D gene (KMT2D) (also known as MLL2), a key regulator of transcriptional enhancer function. KMT2D exhibited truncating nonsense/frameshift/splice site mutations in 8% of SCLC tumors and 17% of SCLC cell lines. We found that KMT2D mutation in human SCLC cell lines was associated with reduced lysine methyltransferase 2D protein levels and reduced monomethylation of histone H3 lysine 4, a mark associated with transcriptional enhancers. We also found mutations in other genes associated with transcriptional enhancer control, including CREB binding protein gene (CREBBP), E1A binding protein p300 gene (EP300), and chromodomain helicase DNA binding protein 7 gene (CHD7), and we report mutations in additional chromatin remodeling genes such as polybromo 1 gene (PBRM1).
Conclusions: These data indicate that KMT2D is one of the major mutated genes in SCLC, and they point to perturbation of transcriptional enhancer control as potentially contributing to SCLC.
Keywords: Genomics; KMT2D; MLL2; SCLC; Small cell lung cancer.
Copyright © 2016 International Association for the Study of Lung Cancer. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure: The authors declare no conflict of interest.
Figures



Comment in
-
Histone Code Aberrancies in Small Cell Lung Cancer.J Thorac Oncol. 2017 Apr;12(4):599-601. doi: 10.1016/j.jtho.2017.02.008. J Thorac Oncol. 2017. PMID: 28343540 No abstract available.
References
-
- Jackman DM, Johnson BE. Small-cell lung cancer. Lancet. 2005;366:1385–1396. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

