Feeding-Related Traits Are Affected by Dosage of the foraging Gene in Drosophila melanogaster
- PMID: 28007892
- PMCID: PMC5289850
- DOI: 10.1534/genetics.116.197939
Feeding-Related Traits Are Affected by Dosage of the foraging Gene in Drosophila melanogaster
Abstract
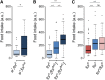
Nutrient acquisition and energy storage are critical parts of achieving metabolic homeostasis. The foraging gene in Drosophila melanogaster has previously been implicated in multiple feeding-related and metabolic traits. Before foraging's functions can be further dissected, we need a precise genetic null mutant to definitively map its amorphic phenotypes. We used homologous recombination to precisely delete foraging, generating the for0 null allele, and used recombineering to reintegrate a full copy of the gene, generating the {forBAC} rescue allele. We show that a total loss of foraging expression in larvae results in reduced larval path length and food intake behavior, while conversely showing an increase in triglyceride levels. Furthermore, varying foraging gene dosage demonstrates a linear dose-response on these phenotypes in relation to foraging gene expression levels. These experiments have unequivocally proven a causal, dose-dependent relationship between the foraging gene and its pleiotropic influence on these feeding-related traits. Our analysis of foraging's transcription start sites, termination sites, and splicing patterns using rapid amplification of cDNA ends (RACE) and full-length cDNA sequencing, revealed four independent promoters, pr1-4, that produce 21 transcripts with nine distinct open reading frames (ORFs). The use of alternative promoters and alternative splicing at the foraging locus creates diversity and flexibility in the regulation of gene expression, and ultimately function. Future studies will exploit these genetic tools to precisely dissect the isoform- and tissue-specific requirements of foraging's functions and shed light on the genetic control of feeding-related traits involved in energy homeostasis.
Keywords: behavior; fat; foraging gene; larva; null mutant.
Copyright © 2017 by the Genetics Society of America.
Figures






References
-
- Anreiter I., Vasquez O. E., Allen A. M., Sokolowski M. B., 2016. Foraging path-length protocol for Drosophila melanogaster larvae. J. Vis. Exp. DOI: 10.3791/53980. - DOI - PMC - PubMed
-
- Bakker K., 1962. An analysis of factors which determine success in competition for food among larvae of Drosophila melanogaster. Arch. Néerl. Zool. 14: 200–281.
-
- Bauer S. J., Sokolowski M. B., 1984. Larval foraging behavior in isofemale lines of Drosophila melanogaster and D. pseudoobscura. J. Hered. 75: 131–134.
-
- Belay A. T., Scheiner R., So A. K.-C., Douglas S. J., Chakaborty-Chatterjee M., et al. , 2007. The foraging gene of Drosophila melanogaster: spatial-expression analysis and sucrose responsiveness. J. Comp. Neurol. 504: 570–582. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

