Frequency doubling in the cyanobacterial circadian clock
- PMID: 28007935
- PMCID: PMC5199125
- DOI: 10.15252/msb.20167087
Frequency doubling in the cyanobacterial circadian clock
Abstract
Organisms use circadian clocks to generate 24-h rhythms in gene expression. However, the clock can interact with other pathways to generate shorter period oscillations. It remains unclear how these different frequencies are generated. Here, we examine this problem by studying the coupling of the clock to the alternative sigma factor sigC in the cyanobacterium Synechococcus elongatus Using single-cell microscopy, we find that psbAI, a key photosynthesis gene regulated by both sigC and the clock, is activated with two peaks of gene expression every circadian cycle under constant low light. This two-peak oscillation is dependent on sigC, without which psbAI rhythms revert to one oscillatory peak per day. We also observe two circadian peaks of elongation rate, which are dependent on sigC, suggesting a role for the frequency doubling in modulating growth. We propose that the two-peak rhythm in psbAI expression is generated by an incoherent feedforward loop between the clock, sigC and psbAI Modelling and experiments suggest that this could be a general network motif to allow frequency doubling of outputs.
Keywords: circadian clock; cyanobacteria; mathematical modelling; network motifs; single‐cell time‐lapse microscopy.
© 2016 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
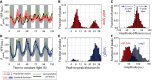
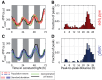
Time traces of PpsbAI‐YFP reporter grown under constant low light (ca. 15 μE m−2 s−1 cool white light). Individual lineages (black and green lines) show the existence of a second peak following the first (dusk timed) peak of expression. Owing to cell‐to‐cell desynchronisation, this second peak is often hidden when expression is measured at the population level (dashed red line, with pink shades representing one standard deviation from the mean). The white and grey shades represent subjective day and subjective night, respectively. 1,319 cells from 10 movies (with up to 419 cells per time point) were collected.
Measure of the distance between the first peak in each circadian cycle and the following peak. The majority of peak pairs occur in the same circadian cycle, with a mean peak‐to‐peak distance of ca. 9 h within this subpopulation.
Distribution of the difference in peak amplitudes between the first and second peaks in a circadian cycle (for cycles where a double peak is present). The second peak is, on average, smaller in amplitude than the first peak.
Neither single‐cell traces nor a population average of psbAI expression shows a double peak in a sigC deletion strain (lines and shades as in A). 1,088 cells from eight movies (with up to 419 cells per time point) were collected.
Measure of the distance between the first peak in each circadian cycle and the following peak shows that the vast majority of lineages have one peak of expression per day.
Distribution of peak amplitudes in wild‐type and sigC deletion strains shows that sigC negatively regulates psbAI expression.
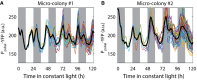
Time traces of PpsbAI‐YFP reporter strains grown under low light (ca. 15 μE m−2 s−1 cool white light). Individual lineages show the existence of a secondary peak following the main (dusk timed) peak of expression. Cells remain synchronised, allowing double peak to be observed in the mean trace (black line).
In a different movie, due to desynchronisation, the double peak is less apparent in the mean trace (black line).
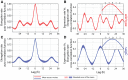
- A
Mean auto‐correlation function of the elongation rate from nine movies (we removed one movie from the analysis as it ended after just 80 h) of the strain carrying the PpsbAI‐YFP reporter in a wild‐type background shows daily double peaks.
- B
Mean cross‐correlation function between expression rate of PpsbAI‐YFP and elongation rate shows daily double peaks and a ca. 1‐h delay of the elongation rate relative to the expression rate (inset with zoom‐in of the mean trace between lags of −3 h and 3 h).
- C, D
Both the mean auto‐correlation function of the elongation rate (C) and the cross‐correlation function between expression rate and elongation rate (D) from eight movies in a sigC deletion background show a single peak per circadian cycle. However, the elongation rate has a similar delay of ca. 1 h relative to the expression rate (D, inset).
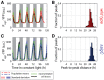
Neither single‐cell traces nor a population average of a reporter for sigC, PsigC‐YFP, shows a double peak of expression. 664 cells from nine movies (with up to 213 cells per time point) were collected.
Measure of the distance between the first peak in each circadian cycle and the following peak confirms that there is only one peak per circadian cycle.
The amplitude in sigC expression is raised by 3.5‐fold in a sigC knock‐out strain, consistent with a sigC‐negative auto‐regulation. 1,084 cells from six movies (with up to 352 cells per time point) were collected.
Measurement of the distance between the first peak in each circadian cycle and the next peak confirms that there is only one peak per circadian cycle.

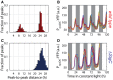
Histogram of PsigC‐YFP expression in wild‐type (red) and sigC deletion mutant (blue) under ca. 15 μE m−2 s−1 cool white light. For the wild type, 664 cells from nine movies were collected, whereas for the sigC deletion strain, 1,084 cells from six movies were collected.
Histogram of PsigC‐YFP expression in wild‐type (red) and sigC deletion mutant (blue) under ca. 35 μE m−2 s−1 cool white light. PsigC‐YFP in wild type is upregulated by twofold between light conditions. A smaller fold change of 1.2 is seen in the sigC deletion mutant. In the higher light condition, 1,003 cells from two movies were collected for the wild‐type strain, whereas 920 cells from two movies were collected for the sigC deletion strain.

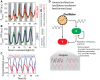
Schematics of the proposed regulatory network. The circadian clock regulates both PsbAI and SigC, and SigC represses both PsbAI and itself. Since the circadian clock regulates PsbAI through two separate branches of opposite signs, one of which is mediated by an intermediate gene, this network contains an incoherent feedforward loop motif.
Numerical simulations of the wild‐type network show double peaks of expression (red line), and numerical simulations of a SigC knock‐out model (in which the terms representing the regulation of PsbAI by SigC are set to zero) show only single‐peaked oscillations (blue line).
The trajectory of the normalised production rate of PsbAI in the wild‐type (red line) simulation shows the dependence of the double peak on the states of the clock and SigC. The surface represents the two‐input production rate function. When the levels of both the clock and SigC are low, the production rate is also low (time point 1, brown circle). As the state of the clock rises, the production rate follows and reaches a plateau (time point 2, which corresponds to the main peak of PsbAI in panel B). However, SigC also rises, crossing a threshold and overcoming the clock, thus imposing a trough in the production rate (time point 3). Later, SigC drops low enough to relieve its negative regulatory activity, and the production rate reaches a second peak (time point 4).
In the SigC deletion simulation (the terms representing the regulation of PsbAI by SigC are set to zero, but SigC expression is tracked for reference), the production rate of PsbAI only responds to the clock, which is a single‐peak oscillation, and so the trajectory of the production rate (blue line) only has a single peak itself (the plateau where time points 2, 3 and 4 lie).
Time traces of PpsbAI‐YFP reporter grown under ca. 35 μE m−2 s−1 cool white light. Most individual lineages (black and green lines) show one circadian peak of gene expression. 2,601 cells from eight movies (with up to 776 cells per time point) were collected.
Measure of the distance between the first peak in each circadian cycle and the following peak shows a single peak per day is the dominant mode.
Time traces of PpsbAI‐YFP reporter in a sigC deletion background look similar to the wild type. 2,306 cells from five movies (with up to 623 cells per time point) were collected.
Measure of peak‐to‐peak distance shows a single circadian peak.

Increasing the deactivation rate of SigC by two orders of magnitude produces only a minor double peak in PsbAI expression. This is comparable to that observed experimentally in higher light conditions (Fig 5A).
The reduction in SigC activity in simulations results in an increase in SigC expression, which qualitatively matches experimental results (Fig EV2). The traces represent the sum of the concentration of the two forms of SigC we considered in the model (see Section II in Appendix).
Time traces of PrpoD6‐YFP reporter grown under low light conditions (ca. 15 μE m−2 s−1 cool white light). Individual lineages (black and green lines) show the existence of a shoulder or a secondary peak of reduced prominence when compared to PpsbAI‐YFP. 947 cells from 12 movies (with up to 272 cells per time point) were collected. Pink shades represent one standard deviation from the mean.
Time traces of PrpoD6‐YFP reporter in a sigC deletion background show only a smooth single‐peaked oscillation. 1,352 cells from seven movies (with up to 502 cells per time point) were collected. Light blue shades represent one standard deviation from the mean.
Numerical simulations of the RpoD6 wild‐type network show a shoulder of expression trailing the main peak (red line). All the parameters describing the clock and SigC are as in Fig 4B, and only the threshold of activation of the rpoD6 promoter by the clock was modified. Numerical simulations of a SigC knock‐out model (in which the terms representing the regulation of RpoD6 by SigC are set to zero) show only single‐peaked oscillations (blue line).
The incoherent feedforward loop circuit that regulates rpoD6 and psbAI is capable of generating diverse oscillatory dynamics in vivo and in silico. Networks where a target gene (Y) is co‐regulated by an oscillator and another regulator (X, which itself is regulated by the oscillator) may represent a general mechanism for generating multi‐peak oscillations.
Measure of the distance between the first peak in each circadian cycle and the following peak shows only a subfraction of cells display a double peak. 947 cells from 12 movies (with up to 272 cells per time point) were collected.
Single‐cell lineages of a representative movie show that all lineages show either a second peak, or a shoulderlike feature, in PrpoD6‐YFP levels.
In the sigC deletion mutant, the two‐peak oscillations are abolished. 1,352 cells from seven movies (with up to 502 cells per time point) were collected.
Single‐cell lineages of a representative movie show a single peak of PrpoD6‐YFP.
Similar articles
-
The cyanobacterial circadian clock couples to pulsatile processes using pulse amplitude modulation.Curr Biol. 2024 Dec 16;34(24):5796-5803.e6. doi: 10.1016/j.cub.2024.10.047. Epub 2024 Nov 25. Curr Biol. 2024. PMID: 39591971
-
Roles for sigma factors in global circadian regulation of the cyanobacterial genome.J Bacteriol. 2002 Jul;184(13):3530-8. doi: 10.1128/JB.184.13.3530-3538.2002. J Bacteriol. 2002. PMID: 12057947 Free PMC article.
-
Light Wavelength as a Contributory Factor of Environmental Fitness in the Cyanobacterial Circadian Clock.Plant Cell Physiol. 2024 May 30;65(5):798-808. doi: 10.1093/pcp/pcae022. Plant Cell Physiol. 2024. PMID: 38441328
-
Circadian Rhythms in Cyanobacteria.Microbiol Mol Biol Rev. 2015 Dec;79(4):373-85. doi: 10.1128/MMBR.00036-15. Microbiol Mol Biol Rev. 2015. PMID: 26335718 Free PMC article. Review.
-
The itty-bitty time machine genetics of the cyanobacterial circadian clock.Adv Genet. 2011;74:13-53. doi: 10.1016/B978-0-12-387690-4.00002-7. Adv Genet. 2011. PMID: 21924974 Free PMC article. Review.
Cited by
-
A Multi-Layered Study on Harmonic Oscillations in Mammalian Genomics and Proteomics.Int J Mol Sci. 2019 Sep 17;20(18):4585. doi: 10.3390/ijms20184585. Int J Mol Sci. 2019. PMID: 31533246 Free PMC article.
-
Environmental and molecular noise buffering by the cyanobacterial clock in individual cells.Nat Commun. 2025 Apr 15;16(1):3566. doi: 10.1038/s41467-025-58169-8. Nat Commun. 2025. PMID: 40234415 Free PMC article.
-
Thermodynamics contributes to high limonene productivity in cyanobacteria.Metab Eng Commun. 2022 Jan 22;14:e00193. doi: 10.1016/j.mec.2022.e00193. eCollection 2022 Jun. Metab Eng Commun. 2022. PMID: 35145855 Free PMC article.
-
Circadian misalignment on submarines and other non-24-h environments - from research to application.Mil Med Res. 2020 Aug 19;7(1):39. doi: 10.1186/s40779-020-00268-2. Mil Med Res. 2020. PMID: 32814592 Free PMC article. Review.
-
An intrinsic oscillator drives the blood stage cycle of the malaria parasite Plasmodium falciparum.Science. 2020 May 15;368(6492):754-759. doi: 10.1126/science.aba4357. Science. 2020. PMID: 32409472 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

