Interaction of p190A RhoGAP with eIF3A and Other Translation Preinitiation Factors Suggests a Role in Protein Biosynthesis
- PMID: 28007963
- PMCID: PMC5314166
- DOI: 10.1074/jbc.M116.769216
Interaction of p190A RhoGAP with eIF3A and Other Translation Preinitiation Factors Suggests a Role in Protein Biosynthesis
Abstract
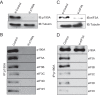
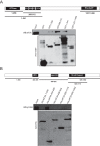
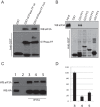
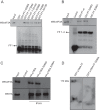
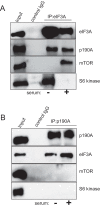
The negative regulator of Rho family GTPases, p190A RhoGAP, is one of six mammalian proteins harboring so-called FF motifs. To explore the function of these and other p190A segments, we identified interacting proteins by tandem mass spectrometry. Here we report that endogenous human p190A, but not its 50% identical p190B paralog, associates with all 13 eIF3 subunits and several other translational preinitiation factors. The interaction involves the first FF motif of p190A and the winged helix/PCI domain of eIF3A, is enhanced by serum stimulation and reduced by phosphatase treatment. The p190A/eIF3A interaction is unaffected by mutating phosphorylated p190A-Tyr308, but disrupted by a S296A mutation, targeting the only other known phosphorylated residue in the first FF domain. The p190A-eIF3 complex is distinct from eIF3 complexes containing S6K1 or mammalian target of rapamycin (mTOR), and appears to represent an incomplete preinitiation complex lacking several subunits. Based on these findings we propose that p190A may affect protein translation by controlling the assembly of functional preinitiation complexes. Whether such a role helps to explain why, unique among the large family of RhoGAPs, p190A exhibits a significantly increased mutation rate in cancer remains to be determined.
Keywords: GTPase activating protein (GAP); Rho (Rho GTPase); cell biology; mass spectrometry (MS); protein biosynthesis; protein complexes; protein domains; translation ini; translation initiation factor; translational initiation.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
-
- Jaffe A. B., and Hall A. (2005) Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21, 247–269 - PubMed
-
- Hall A. (2012) Rho family GTPases. Biochem. Soc. Trans. 40, 1378–1382 - PubMed
-
- Bernards A. (2003) GAPs galore! a survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta 1603, 47–82 - PubMed
-
- Ellis C., Moran M., McCormick F., and Pawson T. (1990) Phosphorylation of GAP and GAP-associated proteins by transforming and mitogenic tyrosine kinases. Nature 343, 377–381 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

