Complex Admixture Preceded and Followed the Extinction of Wisent in the Wild
- PMID: 28007976
- PMCID: PMC5356474
- DOI: 10.1093/molbev/msw254
Complex Admixture Preceded and Followed the Extinction of Wisent in the Wild
Abstract
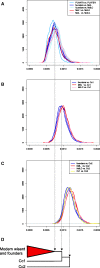
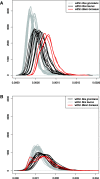
Retracing complex population processes that precede extreme bottlenecks may be impossible using data from living individuals. The wisent (Bison bonasus), Europe's largest terrestrial mammal, exemplifies such a population history, having gone extinct in the wild but subsequently restored by captive breeding efforts. Using low coverage genomic data from modern and historical individuals, we investigate population processes occurring before and after this extinction. Analysis of aligned genomes supports the division of wisent into two previously recognized subspecies, but almost half of the genomic alignment contradicts this population history as a result of incomplete lineage sorting and admixture. Admixture between subspecies populations occurred prior to extinction and subsequently during the captive breeding program. Admixture with the Bos cattle lineage is also widespread but results from ancient events rather than recent hybridization with domestics. Our study demonstrates the huge potential of historical genomes for both studying evolutionary histories and for guiding conservation strategies.
Keywords: Bison bonasus; European bison; admixture; ancient DNA; aurochs; domestic cattle; hybridization..
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
-
- Altizer S, Nunn CL, Lindenfors P. 2007. Do threatened hosts have fewer parasites? A comparative study in primates. J Anim Ecol. 76:304–314. - PubMed
-
- Babik W, Kawałko A, Wójcik JM, Radwan J. 2012. Low major histocompatibility complex class I (MHC I) variation in the European bison (Bison bonasus). J Hered. 349–359. - PubMed
-
- Basrur PK. 1968. Hybrid sterility In: Benirschke K, editor. Comparative Mammalian Cytogenetics. Springer, New York: p. 107–131.
-
- Benecke N. 2005. The Holocene distribution of European bison—the archaeozoological record. Munibe 57:421–428.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

