Microbial competition in porous environments can select against rapid biofilm growth
- PMID: 28007984
- PMCID: PMC5240682
- DOI: 10.1073/pnas.1525228113
Microbial competition in porous environments can select against rapid biofilm growth
Abstract
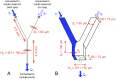
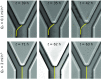
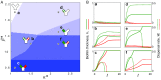
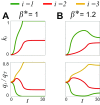
Microbes often live in dense communities called biofilms, where competition between strains and species is fundamental to both evolution and community function. Although biofilms are commonly found in soil-like porous environments, the study of microbial interactions has largely focused on biofilms growing on flat, planar surfaces. Here, we use microfluidic experiments, mechanistic models, and game theory to study how porous media hydrodynamics can mediate competition between bacterial genotypes. Our experiments reveal a fundamental challenge faced by microbial strains that live in porous environments: cells that rapidly form biofilms tend to block their access to fluid flow and redirect resources to competitors. To understand how these dynamics influence the evolution of bacterial growth rates, we couple a model of flow-biofilm interaction with a game theory analysis. This investigation revealed that hydrodynamic interactions between competing genotypes give rise to an evolutionarily stable growth rate that stands in stark contrast with that observed in typical laboratory experiments: cells within a biofilm can outcompete other genotypes by growing more slowly. Our work reveals that hydrodynamics can profoundly affect how bacteria compete and evolve in porous environments, the habitat where most bacteria live.
Keywords: adaptive dynamics; bacterial evolution; clogging; game theory; porous media flow.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



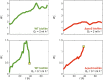
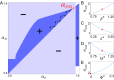
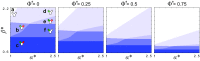
Comment in
-
Reply to Baveye and Darnault: Useful models are simple and extendable.Proc Natl Acad Sci U S A. 2017 Apr 4;114(14):E2804-E2805. doi: 10.1073/pnas.1702303114. Epub 2017 Mar 24. Proc Natl Acad Sci U S A. 2017. PMID: 28341710 Free PMC article. No abstract available.
-
Microbial competition and evolution in natural porous environments: Not that simple.Proc Natl Acad Sci U S A. 2017 Apr 4;114(14):E2802-E2803. doi: 10.1073/pnas.1700992114. Epub 2017 Mar 24. Proc Natl Acad Sci U S A. 2017. PMID: 28341813 Free PMC article. No abstract available.
References
-
- Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5(10):782–791. - PubMed
-
- Bever JD, Richardson SC, Lawrence BM, Holmes J, Watson M. Preferential allocation to beneficial symbiont with spatial structure maintains mycorrhizal mutualism. Ecol Lett. 2009;12(1):13–21. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

