Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding
- PMID: 28008928
- PMCID: PMC5223232
- DOI: 10.1038/cr.2016.152
Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding
Abstract
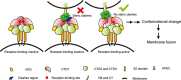
The global outbreak of SARS in 2002-2003 was caused by the infection of a new human coronavirus SARS-CoV. The infection of SARS-CoV is mediated mainly through the viral surface glycoproteins, which consist of S1 and S2 subunits and form trimer spikes on the envelope of the virions. Here we report the ectodomain structures of the SARS-CoV surface spike trimer in different conformational states determined by single-particle cryo-electron microscopy. The conformation 1 determined at 4.3 Å resolution is three-fold symmetric and has all the three receptor-binding C-terminal domain 1 (CTD1s) of the S1 subunits in "down" positions. The binding of the "down" CTD1s to the SARS-CoV receptor ACE2 is not possible due to steric clashes, suggesting that the conformation 1 represents a receptor-binding inactive state. Conformations 2-4 determined at 7.3, 5.7 and 6.8 Å resolutions are all asymmetric, in which one RBD rotates away from the "down" position by different angles to an "up" position. The "up" CTD1 exposes the receptor-binding site for ACE2 engagement, suggesting that the conformations 2-4 represent a receptor-binding active state. This conformational change is also required for the binding of SARS-CoV neutralizing antibodies targeting the CTD1. This phenomenon could be extended to other betacoronaviruses utilizing CTD1 of the S1 subunit for receptor binding, which provides new insights into the intermediate states of coronavirus pre-fusion spike trimer during infection.
Figures




References
-
- Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003; 348:1953–1966. - PubMed
-
- Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003; 348:1967–1976. - PubMed
-
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012; 367:1814–1820. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

