Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis
- PMID: 28009073
- PMCID: PMC5344776
- DOI: 10.1111/mmi.13606
Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis
Abstract
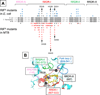
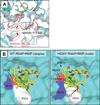
Since 1967, Rifampin (RMP, a Rifamycin) has been used as a first line antibiotic treatment for tuberculosis (TB), and it remains the cornerstone of current short-term TB treatment. Increased occurrence of Rifamycin-resistant (RIFR ) TB, ∼41% of which results from the RpoB S531L mutation in RNA polymerase (RNAP), has become a growing problem worldwide. In this study, we determined the X-ray crystal structures of the Escherichia coli RNAPs containing the most clinically important S531L mutation and two other frequently observed RIFR mutants, RpoB D516V and RpoB H526Y. The structures reveal that the S531L mutation imparts subtle if any structural or functional impact on RNAP in the absence of RIF. However, upon RMP binding, the S531L mutant exhibits a disordering of the RIF binding interface, which effectively reduces the RMP affinity. In contrast, the H526Y mutation reshapes the RIF binding pocket, generating significant steric conflicts that essentially prevent any RIF binding. While the D516V mutant does not exhibit any such gross structural changes, certainly the electrostatic surface of the RIF binding pocket is dramatically changed, likely resulting in the decreased affinity for RIFs. Analysis of interactions of RMP with three common RIFR mutant RNAPs suggests that modifications to RMP may recover its efficacy against RIFR TB.
© 2016 John Wiley & Sons Ltd.
Figures



References
-
- Alekshun M, Kashlev M, Schwartz I. Molecular cloning and characterization of Borrelia burgdorferi rpoB. Gene. 1997;186:227–235. - PubMed
-
- Aristoff PA, Garcia GA, Kirchhoff PD, Showalter HD. Rifamycins - Obstacles and Opportunities. Tuberculosis. 2010;90:94–118. - PubMed
-
- Artsimovitch I, Vassylyeva MN, Svetlov D, Svetlov V, Perederina A, Igarashi N, Matsugaki N, Wakatsuki S, Tahirov TH, Vassylyev DG. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell. 2005;122:351–363. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

