New Monoclonal Antibodies to Defined Cell Surface Proteins on Human Pluripotent Stem Cells
- PMID: 28009074
- PMCID: PMC5412944
- DOI: 10.1002/stem.2558
New Monoclonal Antibodies to Defined Cell Surface Proteins on Human Pluripotent Stem Cells
Abstract
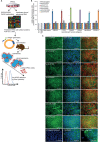
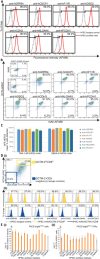
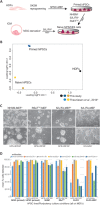
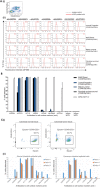
The study and application of human pluripotent stem cells (hPSCs) will be enhanced by the availability of well-characterized monoclonal antibodies (mAbs) detecting cell-surface epitopes. Here, we report generation of seven new mAbs that detect cell surface proteins present on live and fixed human ES cells (hESCs) and human iPS cells (hiPSCs), confirming our previous prediction that these proteins were present on the cell surface of hPSCs. The mAbs all show a high correlation with POU5F1 (OCT4) expression and other hPSC surface markers (TRA-160 and SSEA-4) in hPSC cultures and detect rare OCT4 positive cells in differentiated cell cultures. These mAbs are immunoreactive to cell surface protein epitopes on both primed and naive state hPSCs, providing useful research tools to investigate the cellular mechanisms underlying human pluripotency and states of cellular reprogramming. In addition, we report that subsets of the seven new mAbs are also immunoreactive to human bone marrow-derived mesenchymal stem cells (MSCs), normal human breast subsets and both normal and tumorigenic colorectal cell populations. The mAbs reported here should accelerate the investigation of the nature of pluripotency, and enable development of robust cell separation and tracing technologies to enrich or deplete for hPSCs and other human stem and somatic cell types. Stem Cells 2017;35:626-640.
Keywords: Breast; Cancer; Cell surface markers; Colorectal; Human embryonic stem cells; Human iPS cells; Monoclonal antibodies; Naive; Pluripotency.
© 2016 The Authors Stem Cells published by Wiley Periodicals, Inc. on behalf of AlphaMed Press.
Figures

References
-
- Thomson JA, Itskovitz‐Eldor J, Shapiro SS et al. Embryonic stem cell lines derived from human blastocysts. Science 1998;282:1145–1147. - PubMed
-
- Yu J, Vodyanik MA, Smuga‐Otto K et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007;318:1917–1920. - PubMed
-
- Takahashi K, Tanabe K, Ohnuki M et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861–872. - PubMed

