RNA Structural Determinants of Optimal Codons Revealed by MAGE-Seq
- PMID: 28009265
- PMCID: PMC5234859
- DOI: 10.1016/j.cels.2016.11.004
RNA Structural Determinants of Optimal Codons Revealed by MAGE-Seq
Abstract
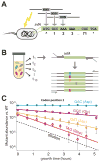
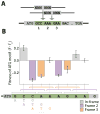
Synonymous codon choices at the beginning of genes optimize 5' RNA structures for enhanced translation initiation, but less is known about mechanisms that drive codon optimization downstream within the gene. To understand what determines codon choices across a gene, we generated 12,726 in situ codon mutants in the Escherichia coli essential gene infA and measured their fitness by combining multiplex automated genome engineering mutagenesis with amplicon deep sequencing (MAGE-seq). Correlating predicted 5' RNA structure with fitness revealed that codons even far from the start of the gene are deleterious if they disrupt the native 5' RNA conformation. These long-range structural interactions generate context-dependent rules that constrain codon choices beyond intrinsic codon preferences. Genome-wide RNA folding predictions confirm that natural codon choices far from the start codon are optimized in part to prevent disruption of native structures near the 5' UTR. Our results shed light on natural codon distributions and should improve engineering of gene expression for synthetic biology applications.
Keywords: RNA structure; codon; codon optimization; codon usage; computational biology; molecular biology; synthetic biology; systems biology.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Codon Clarity or Conundrum?Cell Syst. 2017 Jan 25;4(1):16-19. doi: 10.1016/j.cels.2017.01.004. Cell Syst. 2017. PMID: 28125789 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

