Adenovirus Protein E4-ORF1 Activation of PI3 Kinase Reveals Differential Regulation of Downstream Effector Pathways in Adipocytes
- PMID: 28009298
- PMCID: PMC5193245
- DOI: 10.1016/j.celrep.2016.11.082
Adenovirus Protein E4-ORF1 Activation of PI3 Kinase Reveals Differential Regulation of Downstream Effector Pathways in Adipocytes
Abstract
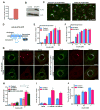
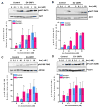
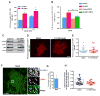
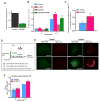
Insulin activation of phosphatidylinositol 3-kinase (PI3K) regulates metabolism, including the translocation of the Glut4 glucose transporter to the plasma membrane and inactivation of the FoxO1 transcription factor. Adenoviral protein E4-ORF1 stimulates cellular glucose metabolism by mimicking growth-factor activation of PI3K. We have used E4-ORF1 as a tool to dissect PI3K-mediated signaling in adipocytes. E4-ORF1 activation of PI3K in adipocytes recapitulates insulin regulation of FoxO1 but not regulation of Glut4. This uncoupling of PI3K effects occurs despite E4-ORF1 activating PI3K and downstream signaling to levels achieved by insulin. Although E4-ORF1 does not fully recapitulate insulin's effects on Glut4, it enhances insulin-stimulated insertion of Glut4-containing vesicles to the plasma membrane independent of Rab10, a key regulator of Glut4 trafficking. E4-ORF1 also stimulates plasma membrane translocation of ubiquitously expressed Glut1 glucose transporter, an effect that is likely essential for E4-ORF1 to promote an anabolic metabolism in a broad range of cell types.
Keywords: E4ORF1; GLUT1; GLUT4; PI3-kinase; PI3K-AKT signaling; RAB10.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Adenovirus E4-ORF1 Dysregulates Epidermal Growth Factor and Insulin/Insulin-Like Growth Factor Receptors To Mediate Constitutive Myc Expression.J Virol. 2015 Nov;89(21):10774-85. doi: 10.1128/JVI.01463-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269183 Free PMC article.
-
Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane.Cell Metab. 2007 Apr;5(4):293-303. doi: 10.1016/j.cmet.2007.03.001. Cell Metab. 2007. PMID: 17403373
-
Adenovirus type 36 regulates adipose stem cell differentiation and glucolipid metabolism through the PI3K/Akt/FoxO1/PPARγ signaling pathway.Lipids Health Dis. 2019 Mar 21;18(1):70. doi: 10.1186/s12944-019-1004-9. Lipids Health Dis. 2019. PMID: 30902099 Free PMC article.
-
Optogenetic activation reveals distinct roles of PIP3 and Akt in adipocyte insulin action.J Cell Sci. 2016 May 15;129(10):2085-95. doi: 10.1242/jcs.174805. Epub 2016 Apr 13. J Cell Sci. 2016. PMID: 27076519 Free PMC article.
-
What we know and what we need to know about adenovirus 36-induced obesity.Int J Obes (Lond). 2020 Jun;44(6):1197-1209. doi: 10.1038/s41366-020-0536-4. Epub 2020 Jan 15. Int J Obes (Lond). 2020. PMID: 31942033 Review.
Cited by
-
Hierarchical regulation of autophagy during adipocyte differentiation.PLoS One. 2022 Jan 26;17(1):e0250865. doi: 10.1371/journal.pone.0250865. eCollection 2022. PLoS One. 2022. PMID: 35081114 Free PMC article.
-
Molecular motion and tridimensional nanoscale localization of kindlin control integrin activation in focal adhesions.Nat Commun. 2021 May 25;12(1):3104. doi: 10.1038/s41467-021-23372-w. Nat Commun. 2021. PMID: 34035280 Free PMC article.
-
A Small Fraction of Progenitors Differentiate Into Mature Adipocytes by Escaping the Constraints on the Cell Structure.Front Cell Dev Biol. 2021 Oct 11;9:753042. doi: 10.3389/fcell.2021.753042. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34708046 Free PMC article.
-
Modelling the gene expression and the DNA-binding in the 3T3-L1 differentiating adipocytes.Adipocyte. 2019 Dec;8(1):401-411. doi: 10.1080/21623945.2019.1697563. Adipocyte. 2019. PMID: 31809632 Free PMC article.
-
Distinct Akt phosphorylation states are required for insulin regulated Glut4 and Glut1-mediated glucose uptake.Elife. 2017 Jun 7;6:e26896. doi: 10.7554/eLife.26896. Elife. 2017. PMID: 28589878 Free PMC article.
References
-
- ABEL ED, PERONI O, KIM JK, KIM YB, BOSS O, HADRO E, MINNEMANN T, SHULMAN GI, KAHN BB. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–33. - PubMed
-
- BAI L, WANG Y, FAN J, CHEN Y, JI W, QU A, XU P, JAMES DE, XU T. Dissecting multiple steps of GLUT4 trafficking and identifying the sites of insulin action. Cell Metab. 2007;5:47–57. - PubMed
-
- BOSE A, GUILHERME A, ROBIDA SI, NICOLORO SM, ZHOU QL, JIANG ZY, POMERLEAU DP, CZECH MP. Glucose transporter recycling in response to insulin is facilitated by myosin Myo1c. Nature. 2002;420:821–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

