Suppression and synthetic-lethal genetic relationships of ΔgpsB mutations indicate that GpsB mediates protein phosphorylation and penicillin-binding protein interactions in Streptococcus pneumoniae D39
- PMID: 28010038
- PMCID: PMC5344783
- DOI: 10.1111/mmi.13613
Suppression and synthetic-lethal genetic relationships of ΔgpsB mutations indicate that GpsB mediates protein phosphorylation and penicillin-binding protein interactions in Streptococcus pneumoniae D39
Abstract
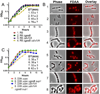
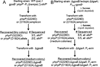
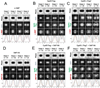
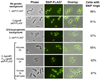
GpsB regulatory protein and StkP protein kinase have been proposed as molecular switches that balance septal and peripheral (side-wall like) peptidoglycan (PG) synthesis in Streptococcus pneumoniae (pneumococcus); yet, mechanisms of this switching remain unknown. We report that ΔdivIVA mutations are not epistatic to ΔgpsB division-protein mutations in progenitor D39 and related genetic backgrounds; nor is GpsB required for StkP localization or FDAA labeling at septal division rings. However, we confirm that reduction of GpsB amount leads to decreased protein phosphorylation by StkP and report that the essentiality of ΔgpsB mutations is suppressed by inactivation of PhpP protein phosphatase, which concomitantly restores protein phosphorylation levels. ΔgpsB mutations are also suppressed by other classes of mutations, including one that eliminates protein phosphorylation and may alter division. Moreover, ΔgpsB mutations are synthetically lethal with Δpbp1a, but not Δpbp2a or Δpbp1b mutations, suggesting GpsB activation of PBP2a activity. Consistent with this result, co-IP experiments showed that GpsB complexes with EzrA, StkP, PBP2a, PBP2b and MreC in pneumococcal cells. Furthermore, depletion of GpsB prevents PBP2x migration to septal centers. These results support a model in which GpsB negatively regulates peripheral PG synthesis by PBP2b and positively regulates septal ring closure through its interactions with StkP-PBP2x.
© 2016 John Wiley & Sons Ltd.
Figures





Comment in
-
The GpsB files: the truth is out there.Mol Microbiol. 2017 Mar;103(6):913-918. doi: 10.1111/mmi.13612. Epub 2017 Jan 12. Mol Microbiol. 2017. PMID: 28010044
Similar articles
-
The Cell Wall of Streptococcus pneumoniae.Microbiol Spectr. 2019 May;7(3):10.1128/microbiolspec.gpp3-0018-2018. doi: 10.1128/microbiolspec.GPP3-0018-2018. Microbiol Spectr. 2019. PMID: 31172911 Free PMC article. Review.
-
Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division.PLoS Genet. 2014 Apr 10;10(4):e1004275. doi: 10.1371/journal.pgen.1004275. eCollection 2014 Apr. PLoS Genet. 2014. PMID: 24722178 Free PMC article.
-
The GpsB files: the truth is out there.Mol Microbiol. 2017 Mar;103(6):913-918. doi: 10.1111/mmi.13612. Epub 2017 Jan 12. Mol Microbiol. 2017. PMID: 28010044
-
Requirement of essential Pbp2x and GpsB for septal ring closure in Streptococcus pneumoniae D39.Mol Microbiol. 2013 Dec;90(5):939-55. doi: 10.1111/mmi.12408. Epub 2013 Oct 17. Mol Microbiol. 2013. PMID: 24118410 Free PMC article.
-
Cell division of Streptococcus pneumoniae: think positive!Curr Opin Microbiol. 2016 Dec;34:18-23. doi: 10.1016/j.mib.2016.07.014. Epub 2016 Aug 4. Curr Opin Microbiol. 2016. PMID: 27497051 Review.
Cited by
-
Lytic transglycosylases: concinnity in concision of the bacterial cell wall.Crit Rev Biochem Mol Biol. 2017 Oct;52(5):503-542. doi: 10.1080/10409238.2017.1337705. Epub 2017 Jun 23. Crit Rev Biochem Mol Biol. 2017. PMID: 28644060 Free PMC article. Review.
-
¡vIVA la DivIVA!J Bacteriol. 2019 Oct 4;201(21):e00245-19. doi: 10.1128/JB.00245-19. Print 2019 Nov 1. J Bacteriol. 2019. PMID: 31405912 Free PMC article. Review.
-
Comparative Phenotypic, Proteomic, and Phosphoproteomic Analysis Reveals Different Roles of Serine/Threonine Phosphatase and Kinase in the Growth, Cell Division, and Pathogenicity of Streptococcus suis.Microorganisms. 2021 Nov 26;9(12):2442. doi: 10.3390/microorganisms9122442. Microorganisms. 2021. PMID: 34946045 Free PMC article.
-
Absence of the KhpA and KhpB (JAG/EloR) RNA-binding proteins suppresses the requirement for PBP2b by overproduction of FtsA in Streptococcus pneumoniae D39.Mol Microbiol. 2017 Dec;106(5):793-814. doi: 10.1111/mmi.13847. Epub 2017 Nov 2. Mol Microbiol. 2017. PMID: 28941257 Free PMC article.
-
The Cell Wall of Streptococcus pneumoniae.Microbiol Spectr. 2019 May;7(3):10.1128/microbiolspec.gpp3-0018-2018. doi: 10.1128/microbiolspec.GPP3-0018-2018. Microbiol Spectr. 2019. PMID: 31172911 Free PMC article. Review.
References
-
- Archambaud C, Gouin E, Pizarro-Cerda J, Cossart P, Dussurget O. Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Molec Microbiol. 2005;56:383–396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

