A feedback mechanism between PLD and deadenylase PARN for the shortening of eukaryotic poly(A) mRNA tails that is deregulated in cancer cells
- PMID: 28011629
- PMCID: PMC5312095
- DOI: 10.1242/bio.021261
A feedback mechanism between PLD and deadenylase PARN for the shortening of eukaryotic poly(A) mRNA tails that is deregulated in cancer cells
Abstract
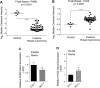
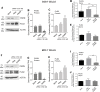
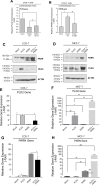
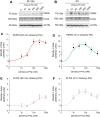
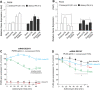
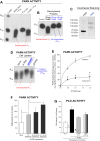
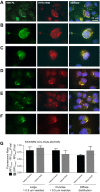
The removal of mRNA transcript poly(A) tails by 3'→5' exonucleases is the rate-limiting step in mRNA decay in eukaryotes. Known cellular deadenylases are the CCR4-NOT and PAN complexes, and poly(A)-specific ribonuclease (PARN). The physiological roles and regulation for PARN is beginning to be elucidated. Since phospholipase D (PLD2 isoform) gene expression is upregulated in breast cancer cells and PARN is downregulated, we examined whether a signaling connection existed between these two enzymes. Silencing PARN with siRNA led to an increase in PLD2 protein, whereas overexpression of PARN had the opposite effect. Overexpression of PLD2, however, led to an increase in PARN expression. Thus, PARN downregulates PLD2 whereas PLD2 upregulates PARN. Co-expression of both PARN and PLD2 mimicked this pattern in non-cancerous cells (COS-7 fibroblasts) but, surprisingly, not in breast cancer MCF-7 cells, where PARN switches from inhibition to activation of PLD2 gene and protein expression. Between 30 and 300 nM phosphatidic acid (PA), the product of PLD enzymatic reaction, added exogenously to culture cells had a stabilizing role of both PARN and PLD2 mRNA decay. Lastly, by immunofluorescence microscopy, we observed an intracellular co-localization of PA-loaded vesicles (0.1-1 nm) and PARN. In summary, we report for the first time the involvement of a phospholipase (PLD2) and PA in mediating PARN-induced eukaryotic mRNA decay and the crosstalk between the two enzymes that is deregulated in breast cancer cells.
Keywords: Breast cancer; Cell signaling; Deadenylase; Gene expression; MRNA decay; Mammalian cells; Ribonuclease.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures

References
-
- Boele J., Persson H., Shin J. W., Ishizu Y., Newie I. S., Sokilde R., Hawkins S. M., Coarfa C., Ikeda K., Takayama K.-i. et al. (2014). PAPD5-mediated 3′ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc. Natl. Acad. Sci. USA 111, 11467-11472. 10.1073/pnas.1317751111 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

