Lipid droplet dynamics during Schizosaccharomyces pombe sporulation and their role in spore survival
- PMID: 28011631
- PMCID: PMC5312105
- DOI: 10.1242/bio.022384
Lipid droplet dynamics during Schizosaccharomyces pombe sporulation and their role in spore survival
Abstract
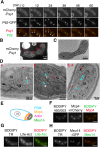
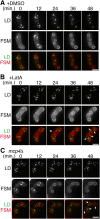
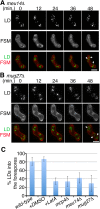
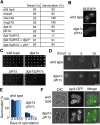
Upon nitrogen starvation, the fission yeast Schizosaccharomyces pombe forms dormant spores; however, the mechanisms by which a spore sustains life without access to exogenous nutrients remain unclear. Lipid droplets are reservoirs of neutral lipids that act as important cellular energy resources. Using live-cell imaging analysis, we found that the lipid droplets of mother cells redistribute to their nascent spores. Notably, this process was actin polymerization-dependent and facilitated by the leading edge proteins of the forespore membrane. Spores lacking triacylglycerol synthesis, which is essential for lipid droplet formation, failed to germinate. Our results suggest that the lipid droplets are important for the sustenance of life in spores.
Keywords: Actin; Forespore membrane; Germination; Lipid droplet; Septation initiation network; Spore.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures
Similar articles
-
Sporulation: A response to starvation in the fission yeast Schizosaccharomyces pombe.Microbiologyopen. 2022 Jun;11(3):e1303. doi: 10.1002/mbo3.1303. Microbiologyopen. 2022. PMID: 35765188 Free PMC article. Review.
-
Meiotic actin rings are essential for proper sporulation in fission yeast.J Cell Sci. 2012 Mar 15;125(Pt 6):1429-39. doi: 10.1242/jcs.091561. J Cell Sci. 2012. PMID: 22526418
-
The protein and neutral lipid composition of lipid droplets isolated from the fission yeast, Schizosaccharomyces pombe.J Microbiol. 2017 Feb;55(2):112-122. doi: 10.1007/s12275-017-6205-1. Epub 2017 Jan 26. J Microbiol. 2017. PMID: 28120187
-
Role for trehalase during germination of spores in the fission yeast Schizosaccharomyces pombe.FEMS Microbiol Lett. 2000 Dec 1;193(1):117-21. doi: 10.1111/j.1574-6968.2000.tb09412.x. FEMS Microbiol Lett. 2000. PMID: 11094289
-
Metabolism of Storage Lipids and the Role of Lipid Droplets in the Yeast Schizosaccharomyces pombe.Lipids. 2020 Sep;55(5):513-535. doi: 10.1002/lipd.12275. Epub 2020 Sep 15. Lipids. 2020. PMID: 32930427 Review.
Cited by
-
Sporulation: A response to starvation in the fission yeast Schizosaccharomyces pombe.Microbiologyopen. 2022 Jun;11(3):e1303. doi: 10.1002/mbo3.1303. Microbiologyopen. 2022. PMID: 35765188 Free PMC article. Review.
-
Molecular and Biophysical Perspectives on Dormancy Breaking: Lessons from Yeast Spore.Biomolecules. 2025 May 11;15(5):701. doi: 10.3390/biom15050701. Biomolecules. 2025. PMID: 40427594 Free PMC article. Review.
-
Lipid droplet motility and organelle contacts.Contact (Thousand Oaks). 2019 Jan-Dec;2:10.1177/2515256419895688. doi: 10.1177/2515256419895688. Epub 2019 Dec 16. Contact (Thousand Oaks). 2019. PMID: 31909374 Free PMC article.
-
Motility Plays an Important Role in the Lifetime of Mammalian Lipid Droplets.Int J Mol Sci. 2021 Apr 7;22(8):3802. doi: 10.3390/ijms22083802. Int J Mol Sci. 2021. PMID: 33916886 Free PMC article. Review.
-
Recovery of Fungal Cells from Air Samples: a Tale of Loss and Gain.Appl Environ Microbiol. 2019 Apr 18;85(9):e02941-18. doi: 10.1128/AEM.02941-18. Print 2019 May 1. Appl Environ Microbiol. 2019. PMID: 30824432 Free PMC article.
References
-
- Bähler J., Wu J.-Q., Longtine M. S., Shah N. G., McKenzie A. III, Steever A. B., Wach A., Philippsen P. and Pringle J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

