Induced Genome-Wide Binding of Three Arabidopsis WRKY Transcription Factors during Early MAMP-Triggered Immunity
- PMID: 28011690
- PMCID: PMC5304350
- DOI: 10.1105/tpc.16.00681
Induced Genome-Wide Binding of Three Arabidopsis WRKY Transcription Factors during Early MAMP-Triggered Immunity
Erratum in
-
CORRECTION.Plant Cell. 2017 May;29(5):1175. doi: 10.1105/tpc.17.00278. Epub 2017 Apr 18. Plant Cell. 2017. PMID: 28420747 Free PMC article. No abstract available.
Abstract
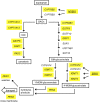
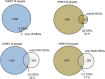
During microbial-associated molecular pattern-triggered immunity (MTI), molecules derived from microbes are perceived by cell surface receptors and upon signaling to the nucleus initiate a massive transcriptional reprogramming critical to mount an appropriate host defense response. WRKY transcription factors play an important role in regulating these transcriptional processes. Here, we determined on a genome-wide scale the flg22-induced in vivo DNA binding dynamics of three of the most prominent WRKY factors, WRKY18, WRKY40, and WRKY33. The three WRKY factors each bound to more than 1000 gene loci predominantly at W-box elements, the known WRKY binding motif. Binding occurred mainly in the 500-bp promoter regions of these genes. Many of the targeted genes are involved in signal perception and transduction not only during MTI but also upon damage-associated molecular pattern-triggered immunity, providing a mechanistic link between these functionally interconnected basal defense pathways. Among the additional targets were genes involved in the production of indolic secondary metabolites and in modulating distinct plant hormone pathways. Importantly, among the targeted genes were numerous transcription factors, encoding predominantly ethylene response factors, active during early MTI, and WRKY factors, supporting the previously hypothesized existence of a WRKY subregulatory network. Transcriptional analysis revealed that WRKY18 and WRKY40 function redundantly as negative regulators of flg22-induced genes often to prevent exaggerated defense responses.
© 2016 American Society of Plant Biologists. All rights reserved.
Figures


References
-
- Albert M. (2013). Peptides as triggers of plant defence. J. Exp. Bot. 64: 5269–5279. - PubMed
-
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152. - PubMed
-
- Assaad F.F., Qiu J.-L., Youngs H., Ehrhardt D., Zimmerli L., Kalde M., Wanner G., Peck S.C., Edwards H., Ramonell K., Somerville C.R., Thordal-Christensen H. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15: 5118–5129. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

