Leukotrienes provide an NFAT-dependent signal that synergizes with IL-33 to activate ILC2s
- PMID: 28011865
- PMCID: PMC5206504
- DOI: 10.1084/jem.20161274
Leukotrienes provide an NFAT-dependent signal that synergizes with IL-33 to activate ILC2s
Abstract
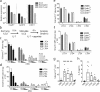
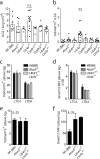
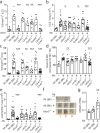
Group 2 innate lymphoid cells (ILC2s) and type 2 helper T cells (Th2 cells) are the primary source of interleukin 5 (IL-5) and IL-13 during type 2 (allergic) inflammation in the lung. In Th2 cells, T cell receptor (TCR) signaling activates the transcription factors nuclear factor of activated T cells (NFAT), nuclear factor κB (NF-κB), and activator protein 1 (AP-1) to induce type 2 cytokines. ILC2s lack a TCR and respond instead to locally produced cytokines such as IL-33. Although IL-33 induces AP-1 and NF-κB, NFAT signaling has not been described in ILC2s. In this study, we report a nonredundant NFAT-dependent role for lipid-derived leukotrienes (LTs) in the activation of lung ILC2s. Using cytokine reporter and LT-deficient mice, we find that complete disruption of LT signaling markedly diminishes ILC2 activation and downstream responses during type 2 inflammation. Type 2 responses are equivalently attenuated in IL-33- and LT-deficient mice, and optimal ILC2 activation reflects potent synergy between these pathways. These findings expand our understanding of ILC2 regulation and may have important implications for the treatment of airways disease.
© 2017 von Moltke et al.
Figures
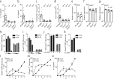

References
-
- Barlow J.L., Bellosi A., Hardman C.S., Drynan L.F., Wong S.H., Cruickshank J.P., and McKenzie A.N.J.. 2012. Innate IL-13–producing nuocytes arise during allergic lung inflammation and contribute to airways hyperreactivity. J. Allergy Clin. Immunol. 129:191–198.e4. 10.1016/j.jaci.2011.09.041 - DOI - PubMed
-
- Barlow J.L., Peel S., Fox J., Panova V., Hardman C.S., Camelo A., Bucks C., Wu X., Kane C.M., Neill D.R., et al. . 2013. IL-33 is more potent than IL-25 in provoking IL-13–producing nuocytes (type 2 innate lymphoid cells) and airway contraction. J. Allergy Clin. Immunol. 132:933–941. 10.1016/j.jaci.2013.05.012 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

