EMC1-dependent stabilization drives membrane penetration of a partially destabilized non-enveloped virus
- PMID: 28012275
- PMCID: PMC5224922
- DOI: 10.7554/eLife.21470
EMC1-dependent stabilization drives membrane penetration of a partially destabilized non-enveloped virus
Abstract
Destabilization of a non-enveloped virus generates a membrane transport-competent viral particle. Here we probe polyomavirus SV40 endoplasmic reticulum (ER)-to-cytosol membrane transport, a decisive infection step where destabilization initiates this non-enveloped virus for membrane penetration. We find that a member of the ER membrane protein complex (EMC) called EMC1 promotes SV40 ER membrane transport and infection. Surprisingly, EMC1 does so by using its predicted transmembrane residue D961 to bind to and stabilize the membrane-embedded partially destabilized SV40, thereby preventing premature viral disassembly. EMC1-dependent stabilization enables SV40 to engage a cytosolic extraction complex that ejects the virus into the cytosol. Thus EMC1 acts as a molecular chaperone, bracing the destabilized SV40 in a transport-competent state. Our findings reveal the novel principle that coordinated destabilization-stabilization drives membrane transport of a non-enveloped virus.
Keywords: cell biology; human; infectious disease; membrane transport; microbiology; protein stabilization; viral entry.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures




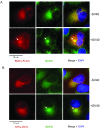
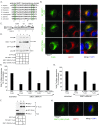


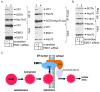
References
-
- Belnap DM, Filman DJ, Trus BL, Cheng N, Booy FP, Conway JF, Curry S, Hiremath CN, Tsang SK, Steven AC, Hogle JM. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. Journal of Virology. 2000;74:1342–1354. doi: 10.1128/JVI.74.3.1342-1354.2000. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

