Autophagy deficient keratinocytes display increased DNA damage, senescence and aberrant lipid composition after oxidative stress in vitro and in vivo
- PMID: 28012437
- PMCID: PMC5192251
- DOI: 10.1016/j.redox.2016.12.015
Autophagy deficient keratinocytes display increased DNA damage, senescence and aberrant lipid composition after oxidative stress in vitro and in vivo
Abstract
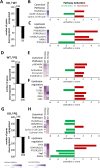
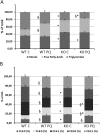
Autophagy allows cells fundamental adaptations to metabolic needs and to stress. Using autophagic bulk degradation cells can clear crosslinked macromolecules and damaged organelles that arise under redox stress. Accumulation of such debris results in cellular dysfunction and is observed in aged tissue and senescent cells. Conversely, promising anti-aging strategies aim at inhibiting the mTOR pathway and thereby activating autophagy, to counteract aging associated damage. We have inactivated autophagy related 7 (Atg7), an essential autophagy gene, in murine keratinocytes (KC) and have found in an earlier study that this resulted in increased baseline oxidative stress and reduced capacity to degrade crosslinked proteins after oxidative ultraviolet stress. To investigate whether autophagy deficiency would promote cellular aging, we studied how Atg7 deficient (KO) and Atg7 bearing cells (WT) would respond to stress induced by paraquat (PQ), an oxidant drug commonly used to induce cellular senescence. Atg7 deficient KC displayed increased prostanoid signaling and a pro- mitotic gene expression signature as compared to the WT. After exposure to PQ, both WT and KO cells showed an inflammatory and stress-related transcriptomic response. However, the Atg7 deficient cells additionally showed drastic DNA damage- and cell cycle arrest signaling. Indeed, DNA fragmentation and -oxidation were strongly increased in the stressed Atg7 deficient cells upon PQ stress but also after oxidizing ultraviolet A irradiation. Damage associated phosphorylated histone H2AX (γH2AX) foci were increased in the nuclei, whereas expression of the nuclear lamina protein lamin B1 was strongly decreased. Similarly, in both, PQ treated mouse tail skin explants and in UVA irradiated mouse tail skin, we found a strong increase in γH2AX positive nuclei within the basal layer of Atg7 deficient epidermis. Atg7 deficiency significantly affected expression of lipid metabolic genes. Therefore we performed lipid profiling of keratinocytes which demonstrated a major dysregulation of cellular lipid metabolism. We found accumulation of autophagy agonisitic free fatty acids, whereas triglyceride levels were strongly decreased. Together, our data show that in absence of Atg7/autophagy the resistance of keratinocytes to intrinsic and environmental oxidative stress was severely impaired and resulted in DNA damage, cell cycle arrest and a disturbed lipid phenotype, all typical for premature cell aging.
Copyright © 2016 The Authors. Published by Elsevier B.V. All rights reserved.
Figures





References
-
- Schafer M., Werner S. Nrf2--A regulator of keratinocyte redox signaling. Free Radic. Biol. Med. 2015;88:243–252. - PubMed
-
- Kammeyer A., Luiten R.M. Oxidation events and skin aging. Ageing Res. Rev. 2015;21:16–29. - PubMed
-
- Latonen L., Laiho M. Cellular UV damage responses--functions of tumor suppressor p53. Biochim. Biophys. Acta. 2005;1755:71–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

