Selective evolution of Toll-like receptors 3, 7, 8, and 9 in bats
- PMID: 28013457
- PMCID: PMC7079974
- DOI: 10.1007/s00251-016-0966-2
Selective evolution of Toll-like receptors 3, 7, 8, and 9 in bats
Abstract
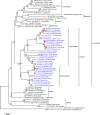
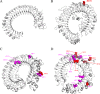
Previous studies have shown that bats are reservoirs of a large number of viruses, many of which cause illness and mortality in humans and other animals. However, these bat-associated pathogens cause little, if any, clinicopathology in bats. This long-term adaptation should be reflected somewhat in the immune system. Toll-like receptors (TLRs) are the first line of immune defense against pathogens in vertebrates. Therefore, this study focuses on the selection of TLRs involved in virus recognition. The coding sequences of TLR3, TLR7, TLR8, and TLR9 were sequenced in ten bats. The selection pressure acting on each gene was also detected using branch- and site-specific methods. The results showed that the ancestor of bats and certain other bat sublineages evolved under positive selection for TLR7, TLR8, and TLR9. The highest proportion of positive selection occurred in TLR9, followed by TLR8 and TLR7. All of the positively selected sites were located in the leucine-rich repeat (LRR) domain, which implied their important roles in pathogen recognition. However, TLR3 evolved under negative selection. Our results are not in line with previous studies which identified more positively selected sites in TLR8 in mammalian species. In this study, the most positively selected sites were found in TLR9. This study encompassed more species that were considered natural reservoirs of viruses. The positive selection for TLR7, TLR8, and TLR9 might contribute to the adaptation of pathogen-host interaction in bats, especially in bat TLR9.
Keywords: Adaptation; Bat; Selective evolution; TLR; Virus defense.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Adaptive evolution of virus-sensing toll-like receptor 8 in bats.Immunogenetics. 2016 Nov;68(10):783-795. doi: 10.1007/s00251-016-0940-z. Epub 2016 Aug 9. Immunogenetics. 2016. PMID: 27502317 Free PMC article.
-
Molecular cloning and expression analysis of toll-like receptor genes (TLR7, TLR8 and TLR9) of golden pompano (Trachinotus ovatus).Fish Shellfish Immunol. 2017 Apr;63:270-276. doi: 10.1016/j.fsi.2017.02.026. Epub 2017 Feb 20. Fish Shellfish Immunol. 2017. PMID: 28232281
-
The functional effects of physical interactions among Toll-like receptors 7, 8, and 9.J Biol Chem. 2006 Dec 8;281(49):37427-34. doi: 10.1074/jbc.M605311200. Epub 2006 Oct 13. J Biol Chem. 2006. PMID: 17040905
-
Human Toll-like receptor-dependent induction of interferons in protective immunity to viruses.Immunol Rev. 2007 Dec;220(1):225-36. doi: 10.1111/j.1600-065X.2007.00564.x. Immunol Rev. 2007. PMID: 17979850 Free PMC article. Review.
-
Regulatory molecules required for nucleotide-sensing Toll-like receptors.Immunol Rev. 2009 Jan;227(1):32-43. doi: 10.1111/j.1600-065X.2008.00729.x. Immunol Rev. 2009. PMID: 19120473 Review.
Cited by
-
Coevolutionary Analysis Implicates Toll-Like Receptor 9 in Papillomavirus Restriction.mBio. 2022 Apr 26;13(2):e0005422. doi: 10.1128/mbio.00054-22. Epub 2022 Mar 21. mBio. 2022. PMID: 35311536 Free PMC article.
-
Processing of genomic RNAs by Dicer in bat cells limits SARS-CoV-2 replication.Virol J. 2025 Mar 25;22(1):86. doi: 10.1186/s12985-025-02693-y. Virol J. 2025. PMID: 40133950 Free PMC article.
-
Bat Red Blood Cells Express Nucleic Acid-Sensing Receptors and Bind RNA and DNA.Immunohorizons. 2022 May 20;6(5):299-306. doi: 10.4049/immunohorizons.2200013. Immunohorizons. 2022. PMID: 35595326 Free PMC article.
-
Antiviral effects of interferon-stimulated genes in bats.Front Cell Infect Microbiol. 2023 Aug 18;13:1224532. doi: 10.3389/fcimb.2023.1224532. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37661999 Free PMC article. Review.
-
Long-Read Sequencing Reveals Rapid Evolution of Immunity- and Cancer-Related Genes in Bats.Genome Biol Evol. 2023 Sep 4;15(9):evad148. doi: 10.1093/gbe/evad148. Genome Biol Evol. 2023. PMID: 37728212 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

