A Syndromic Neurodevelopmental Disorder Caused by De Novo Variants in EBF3
- PMID: 28017372
- PMCID: PMC5223093
- DOI: 10.1016/j.ajhg.2016.11.018
A Syndromic Neurodevelopmental Disorder Caused by De Novo Variants in EBF3
Abstract
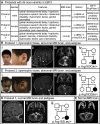
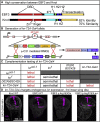
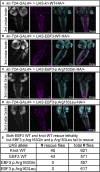
Early B cell factor 3 (EBF3) is a member of the highly evolutionarily conserved Collier/Olf/EBF (COE) family of transcription factors. Prior studies on invertebrate and vertebrate animals have shown that EBF3 homologs are essential for survival and that loss-of-function mutations are associated with a range of nervous system developmental defects, including perturbation of neuronal development and migration. Interestingly, aristaless-related homeobox (ARX), a homeobox-containing transcription factor critical for the regulation of nervous system development, transcriptionally represses EBF3 expression. However, human neurodevelopmental disorders related to EBF3 have not been reported. Here, we describe three individuals who are affected by global developmental delay, intellectual disability, and expressive speech disorder and carry de novo variants in EBF3. Associated features seen in these individuals include congenital hypotonia, structural CNS malformations, ataxia, and genitourinary abnormalities. The de novo variants affect a single conserved residue in a zinc finger motif crucial for DNA binding and are deleterious in a fly model. Our findings indicate that mutations in EBF3 cause a genetic neurodevelopmental syndrome and suggest that loss of EBF3 function might mediate a subset of neurologic phenotypes shared by ARX-related disorders, including intellectual disability, abnormal genitalia, and structural CNS malformations.
Keywords: COE3; Drosophila; ataxia; expressive speech disorder; hypotonia; inhibitory GABAergic neurons; intellectual disability; knot; transcription factor; vermian hypoplasia.
Copyright © 2017 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Alwan A., Modell B. Recommendations for introducing genetics services in developing countries. Nat. Rev. Genet. 2003;4:61–68. - PubMed
-
- American Psychiatric Association, editor. Diagnostic and statistical manual of mental disorders. Fifth Edition. American Psychiatric Association Publishing; 2013.
-
- Boivin M.J., Kakooza A.M., Warf B.C., Davidson L.L., Grigorenko E.L. Reducing neurodevelopmental disorders and disability through research and interventions. Nature. 2015;527:S155–S160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

