FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase
- PMID: 28017614
- PMCID: PMC5234852
- DOI: 10.1016/j.ccell.2016.11.017
FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase
Abstract
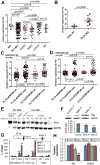
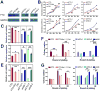
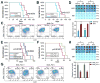
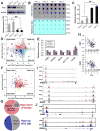
N6-Methyladenosine (m6A) represents the most prevalent internal modification in mammalian mRNAs. Despite its functional importance in various fundamental bioprocesses, the studies of m6A in cancer have been limited. Here we show that FTO, as an m6A demethylase, plays a critical oncogenic role in acute myeloid leukemia (AML). FTO is highly expressed in AMLs with t(11q23)/MLL rearrangements, t(15;17)/PML-RARA, FLT3-ITD, and/or NPM1 mutations. FTO enhances leukemic oncogene-mediated cell transformation and leukemogenesis, and inhibits all-trans-retinoic acid (ATRA)-induced AML cell differentiation, through regulating expression of targets such as ASB2 and RARA by reducing m6A levels in these mRNA transcripts. Collectively, our study demonstrates the functional importance of the m6A methylation and the corresponding proteins in cancer, and provides profound insights into leukemogenesis and drug response.
Keywords: AML; ASB2; ATRA; FTO; RARA; RNA modification; RNA stability; cell differentiation; leukemogenesis; m6A.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




Comment in
-
IDH Mutation, Competitive Inhibition of FTO, and RNA Methylation.Cancer Cell. 2017 May 8;31(5):619-620. doi: 10.1016/j.ccell.2017.04.001. Cancer Cell. 2017. PMID: 28486104 Free PMC article. No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

