Emerging Functions of Circular RNAs
- PMID: 28018143
- PMCID: PMC5168830
Emerging Functions of Circular RNAs
Abstract
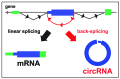
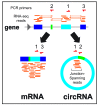
Many thousands of Circular RNAs (circRNAs) have recently been identified in metazoan genomes by transcriptome-wide sequencing. Most circRNAs are generated by back-splicing events from exons of protein-coding genes. A great deal of progress has recently been made in understanding the genome-wide expression patterns, biogenesis, and regulation of circRNAs. To date, however, few functions of circRNAs have been identified. CircRNAs are preferentially expressed in neural tissues and some are found at synapses, suggesting possible functions in the nervous system. Several circRNAs have been shown to function as microRNA "sponges" to counteract microRNA mediated repression of mRNA. New functions for circRNAs are arising, including protein sequestration, transcriptional regulation, and potential functions in cancer. Here, we highlight the recent progress made in understanding the biogenesis and regulation of circRNAs, discuss newly uncovered circRNA functions, and explain the methodological approaches that could reveal more exciting and unexpected roles for these RNAs.
Keywords: EIciRNAs; RNA-seq; aging; alternative splicing; ceRNA; ciRNAs; circRNAs; microRNA; non-coding RNA; transcriptome.
Figures


Similar articles
-
The expanding regulatory mechanisms and cellular functions of circular RNAs.Nat Rev Mol Cell Biol. 2020 Aug;21(8):475-490. doi: 10.1038/s41580-020-0243-y. Epub 2020 May 4. Nat Rev Mol Cell Biol. 2020. PMID: 32366901 Review.
-
The Biogenesis, Functions, and Challenges of Circular RNAs.Mol Cell. 2018 Aug 2;71(3):428-442. doi: 10.1016/j.molcel.2018.06.034. Epub 2018 Jul 26. Mol Cell. 2018. PMID: 30057200 Review.
-
Emerging landscape of circular RNAs in lung cancer.Cancer Lett. 2018 Jul 28;427:18-27. doi: 10.1016/j.canlet.2018.04.006. Epub 2018 Apr 11. Cancer Lett. 2018. PMID: 29653267 Review.
-
Recent progress in circular RNAs in human cancers.Cancer Lett. 2017 Sep 28;404:8-18. doi: 10.1016/j.canlet.2017.07.002. Epub 2017 Jul 11. Cancer Lett. 2017. PMID: 28705773
-
Circular RNA: A new star of noncoding RNAs.Cancer Lett. 2015 Sep 1;365(2):141-8. doi: 10.1016/j.canlet.2015.06.003. Epub 2015 Jun 5. Cancer Lett. 2015. PMID: 26052092 Review.
Cited by
-
Circ-PNPT1 contributes to gestational diabetes mellitus (GDM) by regulating the function of trophoblast cells through miR-889-3p/PAK1 axis.Diabetol Metab Syndr. 2021 Jun 1;13(1):58. doi: 10.1186/s13098-021-00678-9. Diabetol Metab Syndr. 2021. PMID: 34074335 Free PMC article.
-
Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-β signalling.Cell Death Differ. 2020 May;27(5):1709-1727. doi: 10.1038/s41418-019-0455-x. Epub 2019 Nov 14. Cell Death Differ. 2020. PMID: 31728016 Free PMC article.
-
Circular RNAs: Methodological challenges and perspectives in cardiovascular diseases.J Cell Mol Med. 2018 Nov;22(11):5176-5187. doi: 10.1111/jcmm.13789. Epub 2018 Sep 11. J Cell Mol Med. 2018. PMID: 30277664 Free PMC article. Review.
-
Circular RNA 0001823 aggravates the growth and metastasis of the cervical cancer cells through modulating the microRNA-613/RAB8A axis.Bioengineered. 2022 Apr;13(4):10335-10349. doi: 10.1080/21655979.2022.2063665. Bioengineered. 2022. PMID: 35435110 Free PMC article.
-
The investigation of circRNA profiling reveals the regulatory role of the hsa_circ_0000375/miR-7706 pathway in breast cancer.Mol Biol Rep. 2023 Dec;50(12):9993-10004. doi: 10.1007/s11033-023-08798-3. Epub 2023 Oct 30. Mol Biol Rep. 2023. PMID: 37904009
References
-
- Capel B, Swain A, Nicolis S. et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–1030. - PubMed
-
- Nigro JM, Cho KR, Fearon ER. et al. Scrambled exons. Cell. 1991;64(3):607–613. - PubMed
-
- Memczak S, Jens M, Elefsinioti A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
