NKG2A-Expressing Natural Killer Cells Dominate the Response to Autologous Lymphoblastoid Cells Infected with Epstein-Barr Virus
- PMID: 28018364
- PMCID: PMC5156658
- DOI: 10.3389/fimmu.2016.00607
NKG2A-Expressing Natural Killer Cells Dominate the Response to Autologous Lymphoblastoid Cells Infected with Epstein-Barr Virus
Abstract
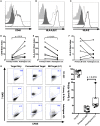
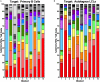
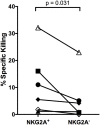
Epstein-Barr virus (EBV) is a human γ-herpesvirus that establishes latency and lifelong infection in host B cells while achieving a balance with the host immune response. When the immune system is perturbed through immunosuppression or immunodeficiency, however, these latently infected B cells can give rise to aggressive B cell lymphomas. Natural killer (NK) cells are regarded as critical in the early immune response to viral infection, but their role in controlling expansion of infected B cells is not understood. Here, we report that NK cells from healthy human donors display increased killing of autologous B lymphoblastoid cell lines (LCLs) harboring latent EBV compared to primary B cells. Coculture of NK cells with autologous EBV+ LCL identifies an NK cell population that produces IFNγ and mobilizes the cytotoxic granule protein CD107a. Multi-parameter flow cytometry and Boolean analysis reveal that these functional cells are enriched for expression of the NK cell receptor NKG2A. Further, NKG2A+ NK cells more efficiently lyse autologous LCL than do NKG2A- NK cells. More specifically, NKG2A+2B4+CD16-CD57-NKG2C-NKG2D+ cells constitute the predominant NK cell population that responds to latently infected autologous EBV+ B cells. Thus, a subset of NK cells is enhanced for the ability to recognize and eliminate autologous, EBV-infected transformed cells, laying the groundwork for harnessing this subset for therapeutic use in EBV+ malignancies.
Keywords: B cells; Epstein–Barr virus; NK cells; NKG2A; lymphoblastoid cell lines.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

