Chloroplast Genome Sequence of Pigeonpea (Cajanus cajan (L.) Millspaugh) and Cajanus scarabaeoides (L.) Thouars: Genome Organization and Comparison with Other Legumes
- PMID: 28018385
- PMCID: PMC5145887
- DOI: 10.3389/fpls.2016.01847
Chloroplast Genome Sequence of Pigeonpea (Cajanus cajan (L.) Millspaugh) and Cajanus scarabaeoides (L.) Thouars: Genome Organization and Comparison with Other Legumes
Abstract
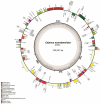
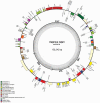
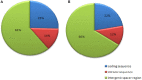
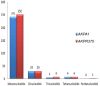
Pigeonpea (Cajanus cajan (L.) Millspaugh), a diploid (2n = 22) legume crop with a genome size of 852 Mbp, serves as an important source of human dietary protein especially in South East Asian and African regions. In this study, the draft chloroplast genomes of Cajanus cajan and Cajanus scarabaeoides (L.) Thouars were generated. Cajanus scarabaeoides is an important species of the Cajanus gene pool and has also been used for developing promising CMS system by different groups. A male sterile genotype harboring the C. scarabaeoides cytoplasm was used for sequencing the plastid genome. The cp genome of C. cajan is 152,242bp long, having a quadripartite structure with LSC of 83,455 bp and SSC of 17,871 bp separated by IRs of 25,398 bp. Similarly, the cp genome of C. scarabaeoides is 152,201bp long, having a quadripartite structure in which IRs of 25,402 bp length separates 83,423 bp of LSC and 17,854 bp of SSC. The pigeonpea cp genome contains 116 unique genes, including 30 tRNA, 4 rRNA, 78 predicted protein coding genes and 5 pseudogenes. A 50 kb inversion was observed in the LSC region of pigeonpea cp genome, consistent with other legumes. Comparison of cp genome with other legumes revealed the contraction of IR boundaries due to the absence of rps19 gene in the IR region. Chloroplast SSRs were mined and a total of 280 and 292 cpSSRs were identified in C. scarabaeoides and C. cajan respectively. RNA editing was observed at 37 sites in both C. scarabaeoides and C. cajan, with maximum occurrence in the ndh genes. The pigeonpea cp genome sequence would be beneficial in providing informative molecular markers which can be utilized for genetic diversity analysis and aid in understanding the plant systematics studies among major grain legumes.
Keywords: Cajanus cajan; Cajanus scarabaeoides; RNA editing; Roche 454 sequencing; chloroplast genome.
Figures



Similar articles
-
Comparative transcriptome analyses revealed different heat stress responses in pigeonpea (Cajanus cajan) and its crop wild relatives.Plant Cell Rep. 2021 May;40(5):881-898. doi: 10.1007/s00299-021-02686-5. Epub 2021 Apr 10. Plant Cell Rep. 2021. PMID: 33837822
-
Development of new cytoplasmic-genetic male-sterile lines in pigeonpea from crosses between Cajanus cajan (L.) Millsp.and C. scarabaeoides (L.) Thouars.J Appl Genet. 2008;49(3):221-7. doi: 10.1007/BF03195617. J Appl Genet. 2008. PMID: 18670057
-
De novo Assembly and Characterization of Cajanus scarabaeoides (L.) Thouars Transcriptome by Paired-End Sequencing.Front Mol Biosci. 2017 Jul 12;4:48. doi: 10.3389/fmolb.2017.00048. eCollection 2017. Front Mol Biosci. 2017. PMID: 28748187 Free PMC article.
-
Pigeonpea (Cajanus cajan L. Millsp.).Methods Mol Biol. 2006;343:359-67. doi: 10.1385/1-59745-130-4:359. Methods Mol Biol. 2006. PMID: 16988359 Review.
-
Revisiting the Nutritional, Chemical and Biological Potential of Cajanus cajan (L.) Millsp.Molecules. 2022 Oct 13;27(20):6877. doi: 10.3390/molecules27206877. Molecules. 2022. PMID: 36296470 Free PMC article. Review.
Cited by
-
New Insights Into the Plastome Evolution of the Millettioid/Phaseoloid Clade (Papilionoideae, Leguminosae).Front Plant Sci. 2020 Mar 10;11:151. doi: 10.3389/fpls.2020.00151. eCollection 2020. Front Plant Sci. 2020. PMID: 32210983 Free PMC article.
-
Uncovering dynamic evolution in the plastid genome of seven Ligusticum species provides insights into species discrimination and phylogenetic implications.Sci Rep. 2021 Jan 13;11(1):988. doi: 10.1038/s41598-020-80225-0. Sci Rep. 2021. PMID: 33441833 Free PMC article.
-
Plastome evolution of Engelhardia facilitates phylogeny of Juglandaceae.BMC Plant Biol. 2024 Jul 6;24(1):634. doi: 10.1186/s12870-024-05293-0. BMC Plant Biol. 2024. PMID: 38971744 Free PMC article.
-
Chloroplast genome sequence of Chongming lima bean (Phaseolus lunatus L.) and comparative analyses with other legume chloroplast genomes.BMC Genomics. 2021 Mar 18;22(1):194. doi: 10.1186/s12864-021-07467-8. BMC Genomics. 2021. PMID: 33736599 Free PMC article.
-
Analysis of the complete plastomes and nuclear ribosomal DNAs from Euonymus hamiltonianus and its relatives sheds light on their diversity and evolution.PLoS One. 2022 Oct 5;17(10):e0275590. doi: 10.1371/journal.pone.0275590. eCollection 2022. PLoS One. 2022. PMID: 36197898 Free PMC article.
References
-
- Bock R. (2007). Structure, function, and inheritance of plastid genomes, in Cell and Molecular Biology of Plastids, ed Bock R.(Berlin; Heidelberg: Springer; ), 29–63.
-
- Bruneau A., Doyle J. J., Palmer J. D. (1990). A Chloroplast DNA Inversion as a subtribal character in the Phaseoleae (Leguminosae). Syst. Bot. 15, 378–386. 10.2307/2419351 - DOI
-
- Bulmer M. (1988). Are codon usage patterns in unicellular organisms determined by selection-mutation balance? J. Evol. Biol. 1, 15–26. 10.1046/j.1420-9101.1988.1010015.x - DOI
-
- Cai Z., Guisinger M., Kim H. G., Ruck E., Blazier J. C., McMurtry V., et al. . (2008). Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J. Mol. Evol. 67, 696–704. 10.1007/s00239-008-9180-7 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

