Identification of Plastoglobules as a Site of Carotenoid Cleavage
- PMID: 28018391
- PMCID: PMC5161054
- DOI: 10.3389/fpls.2016.01855
Identification of Plastoglobules as a Site of Carotenoid Cleavage
Abstract
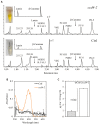
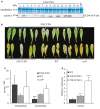
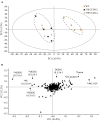
Carotenoids play an essential role in light harvesting and protection from excess light. During chloroplast senescence carotenoids are released from their binding proteins and are eventually metabolized. Carotenoid cleavage dioxygenase 4 (CCD4) is involved in carotenoid breakdown in senescing leaf and desiccating seed, and is part of the proteome of plastoglobules (PG), which are thylakoid-associated lipid droplets. Here, we demonstrate that CCD4 is functionally active in PG. Leaves of Arabidopsis thaliana ccd4 mutants constitutively expressing CCD4 fused to yellow fluorescent protein showed strong fluorescence in PG and reduced carotenoid levels upon dark-induced senescence. Lipidome-wide analysis indicated that β-carotene, lutein, and violaxanthin were the principle substrates of CCD4 in vivo and were cleaved in senescing chloroplasts. Moreover, carotenoids were shown to accumulate in PG of ccd4 mutant plants during senescence, indicating translocation of carotenoids to PG prior to degradation.
Keywords: Arabidopsis thaliana; carotenoid; ccd4; chloroplast; plastoglobule; senescence.
Figures


Similar articles
-
Carotenoid cleavage dioxygenase4 is a negative regulator of β-carotene content in Arabidopsis seeds.Plant Cell. 2013 Dec;25(12):4812-26. doi: 10.1105/tpc.113.119677. Epub 2013 Dec 24. Plant Cell. 2013. PMID: 24368792 Free PMC article.
-
Functions and substrates of plastoglobule-localized metallopeptidase PGM48.Plant Signal Behav. 2017 Jun 3;12(6):e1331197. doi: 10.1080/15592324.2017.1331197. Epub 2017 May 23. Plant Signal Behav. 2017. PMID: 28534654 Free PMC article.
-
Molecular changes of Arabidopsis thaliana plastoglobules facilitate thylakoid membrane remodeling under high light stress.Plant J. 2021 Jun;106(6):1571-1587. doi: 10.1111/tpj.15253. Epub 2021 May 3. Plant J. 2021. PMID: 33783866
-
The role of plastoglobules in thylakoid lipid remodeling during plant development.Biochim Biophys Acta. 2015 Sep;1847(9):889-99. doi: 10.1016/j.bbabio.2015.02.002. Epub 2015 Feb 7. Biochim Biophys Acta. 2015. PMID: 25667966 Review.
-
A mechanism implicating plastoglobules in thylakoid disassembly during senescence and nitrogen starvation.Planta. 2013 Feb;237(2):463-70. doi: 10.1007/s00425-012-1813-9. Epub 2012 Nov 28. Planta. 2013. PMID: 23187680 Review.
Cited by
-
Nutritional Enrichment of Plant Leaves by Combining Genes Promoting Tocopherol Biosynthesis and Storage.Metabolites. 2023 Jan 28;13(2):193. doi: 10.3390/metabo13020193. Metabolites. 2023. PMID: 36837812 Free PMC article.
-
Time Course of Age-Linked Changes in Photosynthetic Efficiency of Spirodela polyrhiza Exposed to Cadmium.Front Plant Sci. 2022 May 25;13:872793. doi: 10.3389/fpls.2022.872793. eCollection 2022. Front Plant Sci. 2022. PMID: 35693160 Free PMC article.
-
Transcriptome Analysis Reveals Candidate Genes Associated with Leaf Etiolation of a Cytoplasmic Male Sterility Line in Chinese Cabbage (Brassica Rapa L. ssp. Pekinensis).Int J Mol Sci. 2018 Mar 21;19(4):922. doi: 10.3390/ijms19040922. Int J Mol Sci. 2018. PMID: 29561749 Free PMC article.
-
Leaf Senescence: The Chloroplast Connection Comes of Age.Plants (Basel). 2019 Nov 12;8(11):495. doi: 10.3390/plants8110495. Plants (Basel). 2019. PMID: 31718069 Free PMC article. Review.
-
Diversity of Plastid Types and Their Interconversions.Front Plant Sci. 2021 Jun 17;12:692024. doi: 10.3389/fpls.2021.692024. eCollection 2021. Front Plant Sci. 2021. PMID: 34220916 Free PMC article. Review.
References
-
- Ariizumi T., Kishimoto S., Kakami R., Maoka T., Hirakawa H., Suzuki Y., et al. (2014). Identification of the carotenoid modifying gene PALE YELLOW PETAL 1 as an essential factor in xanthophyll esterification and yellow flower pigmentation in tomato (Solanum lycopersicum). Plant J. 79 453–465. 10.1111/tpj.12570 - DOI - PubMed
-
- Avendano-Vazquez A. O., Cordoba E., Llamas E., San Roman C., Nisar N., De la Torre S., et al. (2014). An uncharacterized apocarotenoid-derived signal generated in zeta-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis. Plant Cell 26 2524–2537. 10.1105/tpc.114.123349 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

