Molecular Evolution and Expression Divergence of the Aconitase (ACO) Gene Family in Land Plants
- PMID: 28018410
- PMCID: PMC5149538
- DOI: 10.3389/fpls.2016.01879
Molecular Evolution and Expression Divergence of the Aconitase (ACO) Gene Family in Land Plants
Abstract
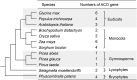
Aconitase (ACO) is a key enzyme that catalyzes the isomerization of citrate to isocitrate in the tricarboxylic acid (TCA) and glyoxylate cycles. The function of ACOs has been well studied in model plants, such as Arabidopsis. In contrast, the evolutionary patterns of the ACO family in land plants are poorly understood. In this study, we systematically examined the molecular evolution and expression divergence of the ACO gene family in 12 land plant species. Thirty-six ACO genes were identified from the 12 land plant species representing the four major land plant lineages: Bryophytes, lycophytes, gymnosperms, and angiosperms. All of these ACOs belong to the cytosolic isoform. Three gene duplication events contributed to the expansion of the ACO family in angiosperms. The ancestor of angiosperms may have contained only one ACO gene. One gene duplication event split angiosperm ACOs into two distinct clades. Two clades showed a divergence in selective pressure and gene expression patterns. The cis-acting elements that function in light responsiveness were most abundant in the promoter region of the ACO genes, indicating that plant ACO genes might participate in light regulatory pathways. Our findings provide comprehensive insights into the ACO gene family in land plants.
Keywords: aconitase; evolution; gene expression; gene family; land plants.
Figures








Similar articles
-
Genome-Wide Identification and Expression Profiling of Aconitase Gene Family Members Reveals Their Roles in Plant Development and Adaptation to Diverse Stress in Triticum aestivum L.Plants (Basel). 2022 Dec 12;11(24):3475. doi: 10.3390/plants11243475. Plants (Basel). 2022. PMID: 36559588 Free PMC article.
-
Conservation and diversification of HAIRY MERISTEM gene family in land plants.Plant J. 2021 Apr;106(2):366-378. doi: 10.1111/tpj.15169. Epub 2021 Mar 27. Plant J. 2021. PMID: 33484592
-
Conservation and divergence of small RNA pathways and microRNAs in land plants.Genome Biol. 2017 Aug 23;18(1):158. doi: 10.1186/s13059-017-1291-2. Genome Biol. 2017. PMID: 28835265 Free PMC article.
-
The major clades of MADS-box genes and their role in the development and evolution of flowering plants.Mol Phylogenet Evol. 2003 Dec;29(3):464-89. doi: 10.1016/s1055-7903(03)00207-0. Mol Phylogenet Evol. 2003. PMID: 14615187 Review.
-
Evolution of Cell Wall Polymers in Tip-Growing Land Plant Gametophytes: Composition, Distribution, Functional Aspects and Their Remodeling.Front Plant Sci. 2019 Apr 18;10:441. doi: 10.3389/fpls.2019.00441. eCollection 2019. Front Plant Sci. 2019. PMID: 31057570 Free PMC article. Review.
Cited by
-
Genome-Wide Identification and Expression Profiling of Aconitase Gene Family Members Reveals Their Roles in Plant Development and Adaptation to Diverse Stress in Triticum aestivum L.Plants (Basel). 2022 Dec 12;11(24):3475. doi: 10.3390/plants11243475. Plants (Basel). 2022. PMID: 36559588 Free PMC article.
-
Transcriptome Analysis of Bread Wheat Genotype KRL3-4 Provides a New Insight Into Regulatory Mechanisms Associated With Sodicity (High pH) Tolerance.Front Genet. 2022 Feb 9;12:782366. doi: 10.3389/fgene.2021.782366. eCollection 2021. Front Genet. 2022. PMID: 35222517 Free PMC article.
-
Transcriptome analysis to identify genes related to programmed cell death resulted from manipulating of BnaFAH ortholog by CRISPR/Cas9 in Brassica napus.Sci Rep. 2024 Nov 2;14(1):26389. doi: 10.1038/s41598-024-77877-7. Sci Rep. 2024. PMID: 39488592 Free PMC article.
-
Aconitase: To Be or not to Be Inside Plant Glyoxysomes, That Is the Question.Biology (Basel). 2020 Jul 12;9(7):162. doi: 10.3390/biology9070162. Biology (Basel). 2020. PMID: 32664680 Free PMC article. Review.
-
A systematic in silico report on iron and zinc proteome of Zea mays.Front Plant Sci. 2023 Aug 17;14:1166720. doi: 10.3389/fpls.2023.1166720. eCollection 2023. Front Plant Sci. 2023. PMID: 37662157 Free PMC article.
References
-
- Carrari F., Nunes-Nesi A., Gibon Y., Lytovchenko A., Loureiro M. E., Fernie A. R. (2003). Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol. 133, 1322–1335. 10.1104/pp.103.026716 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

