Can terminators be used as insulators into yeast synthetic gene circuits?
- PMID: 28018483
- PMCID: PMC5162094
- DOI: 10.1186/s13036-016-0040-5
Can terminators be used as insulators into yeast synthetic gene circuits?
Abstract
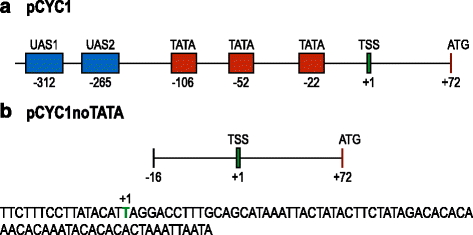
Background: In bacteria, transcription units can be insulated by placing a terminator in front of a promoter. In this way promoter leakage due to the read-through from an upstream gene or RNA polymerase unspecific binding to the DNA is, in principle, removed. Differently from bacterial terminators, yeast S. cerevisiae terminators contain a hexamer sequence, the efficiency element, that strongly resembles the eukaryotic TATA box i.e. the promoter sequence recognized and bound by RNA polymerase II.
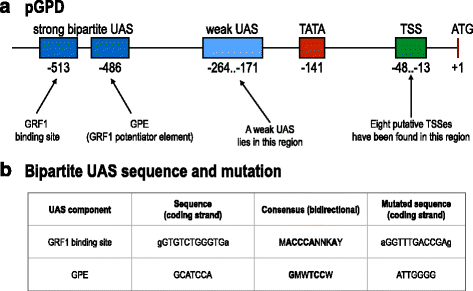
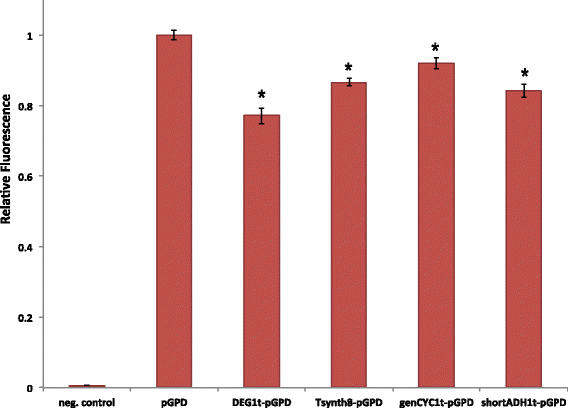
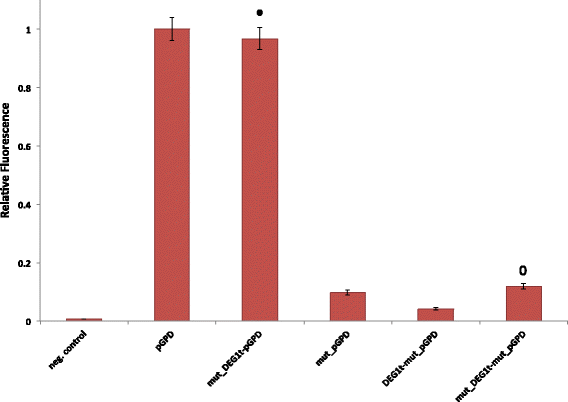
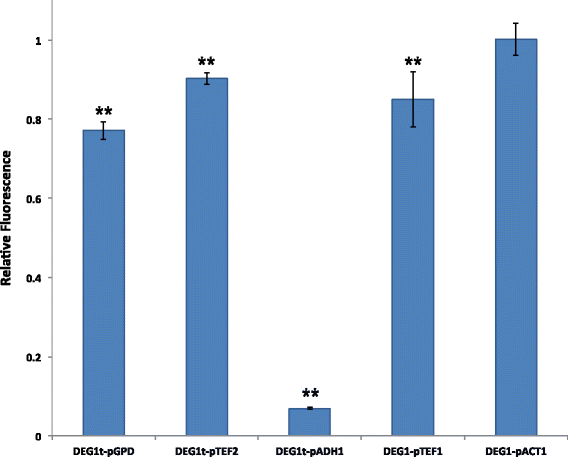
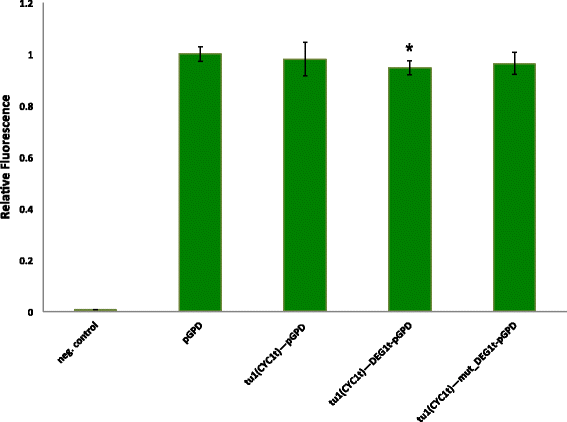
Results: By placing different yeast terminators (natural and synthetic) in front of the CYC1 yeast constitutive promoter stripped of every upstream activating sequences and TATA boxes, we verified that the efficiency element is able to bind RNA polymerase II, hence working as a TATA box. Moreover, terminators put in front of strong and medium-strength constitutive yeast promoters cause a non-negligible decrease in the promoter transcriptional activity.
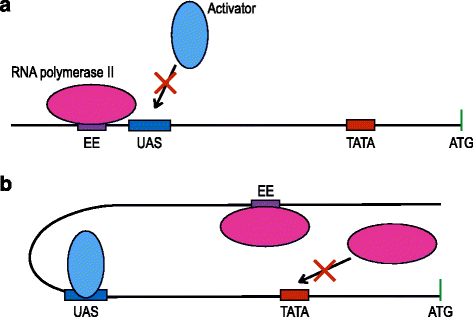
Conclusions: Our data suggests that RNA polymerase II molecules upon binding the insulator efficiency element interfere with protein expression by competing either with activator proteins at the promoter enhancers or other RNA polymerase II molecules targeting the TATA box. Hence, it seems preferable to avoid the insulation of non-weak promoters when building synthetic gene circuit in yeast S. cerevisiae.
Keywords: Efficiency element; Insulation; S. cerevisiae; TATA box; Terminator.
Figures



Similar articles
-
Development of Terminator-Promoter Bifunctional Elements for Application in Saccharomyces cerevisiae Pathway Engineering.Int J Mol Sci. 2023 Jun 7;24(12):9870. doi: 10.3390/ijms24129870. Int J Mol Sci. 2023. PMID: 37373018 Free PMC article.
-
Yeast Synthetic Terminators: Fine Regulation of Strength through Linker Sequences.Chembiochem. 2019 Sep 16;20(18):2383-2389. doi: 10.1002/cbic.201900163. Epub 2019 Jul 8. Chembiochem. 2019. PMID: 30974044
-
Binding of TFIID to the CYC1 TATA boxes in yeast occurs independently of upstream activating sequences.Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11909-13. doi: 10.1073/pnas.91.25.11909. Proc Natl Acad Sci U S A. 1994. PMID: 7991556 Free PMC article.
-
TRF2: TRansForming the view of general transcription factors.Transcription. 2015;6(1):1-6. doi: 10.1080/21541264.2015.1004980. Epub 2015 Jan 14. Transcription. 2015. PMID: 25588059 Free PMC article. Review.
-
The DPE, a core promoter element for transcription by RNA polymerase II.Exp Mol Med. 2002 Sep 30;34(4):259-64. doi: 10.1038/emm.2002.36. Exp Mol Med. 2002. PMID: 12515390 Review.
Cited by
-
Efficient sex hormone biosensors in Saccharomyces cerevisiae cells to evaluate human aromatase activity and inhibition.Sci Rep. 2025 Jan 3;15(1):737. doi: 10.1038/s41598-024-85022-7. Sci Rep. 2025. PMID: 39753751 Free PMC article.
-
Tuning heterologous glucan biosynthesis in yeast to understand and exploit plant starch diversity.BMC Biol. 2022 Sep 24;20(1):207. doi: 10.1186/s12915-022-01408-x. BMC Biol. 2022. PMID: 36153520 Free PMC article.
-
Hybrid Synthetic Promoters in Saccharomyces cerevisiae Built on Foreign Promoter Sequences.Methods Mol Biol. 2024;2844:109-119. doi: 10.1007/978-1-0716-4063-0_7. Methods Mol Biol. 2024. PMID: 39068335
-
Systems and Synthetic Biology Approaches to Engineer Fungi for Fine Chemical Production.Front Bioeng Biotechnol. 2018 Oct 3;6:117. doi: 10.3389/fbioe.2018.00117. eCollection 2018. Front Bioeng Biotechnol. 2018. PMID: 30338257 Free PMC article. Review.
-
Hybrid Boolean gates show that Cas12c controls transcription activation effectively in the yeast S. cerevisiae.Front Bioeng Biotechnol. 2023 Sep 12;11:1267174. doi: 10.3389/fbioe.2023.1267174. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 37771576 Free PMC article.
References
-
- Alon U. An introduction to systems biology. Boca Raton: Chapman & Hall/CRC Press; 2006.
-
- Lewin B. genes VII. New York: Oxford University Press; 2000.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

